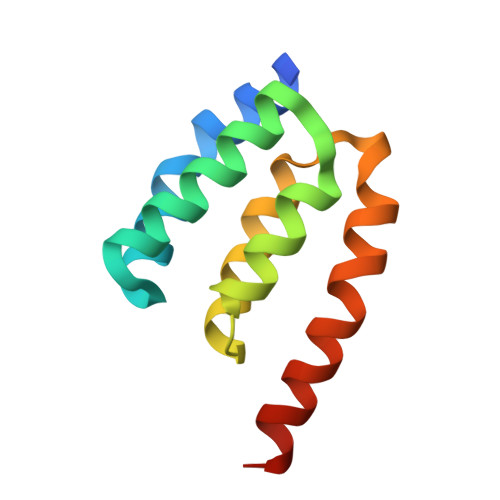
Structure and Membrane Targeting of the PDZD7 Harmonin Homology Domain (HHD) Associated With Hearing Loss.
Lin, L., Wang, H., Ren, D., Xia, Y., He, G., Lu, Q.(2021) Front Cell Dev Biol 9: 642666-642666
- PubMed: 33937240
- DOI: https://doi.org/10.3389/fcell.2021.642666
- Primary Citation of Related Structures:
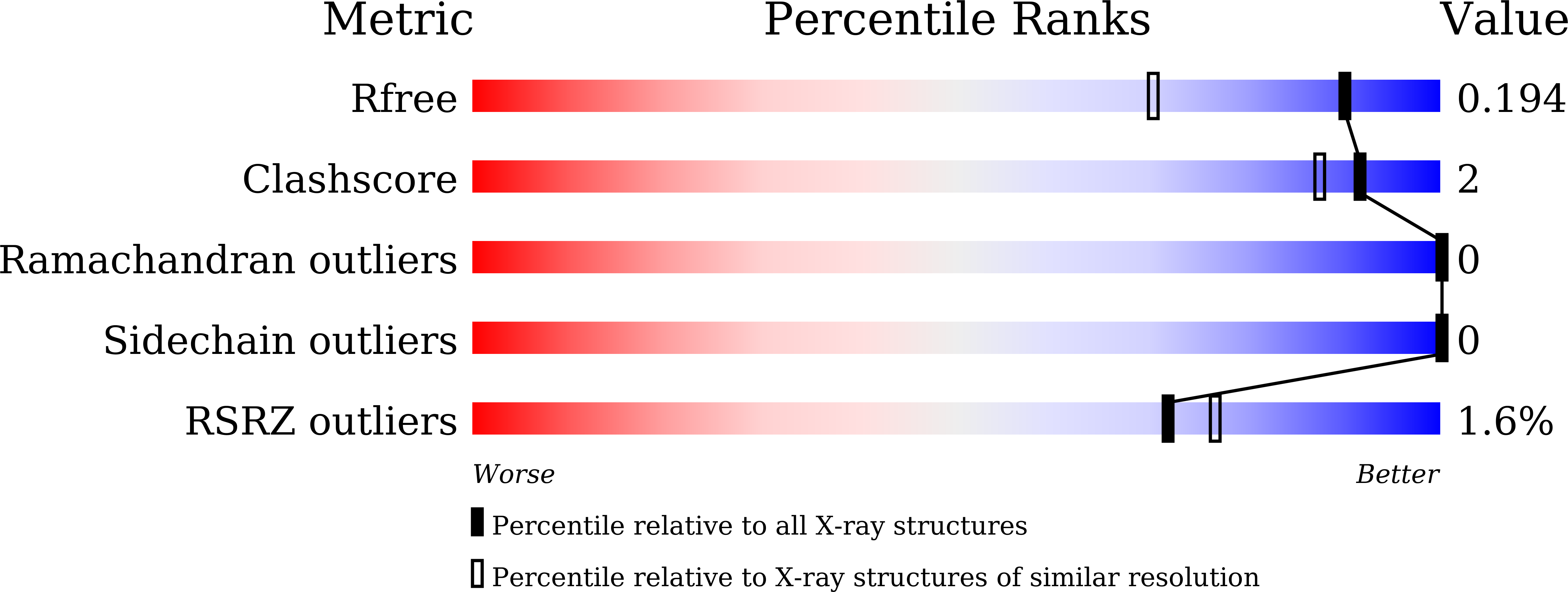
7DE7 - PubMed Abstract:
Usher syndrome (USH) is the leading cause of hereditary hearing-vision loss in humans. PDZ domain-containing 7 (PDZD7) has been reported to be a modifier of and contributor to USH. PDZD7 co-localizes with USH2 proteins in the inner ear hair cells and is essential for ankle-link formation and stereocilia development. PDZD7 contains three PDZ domains and a low-complexity region between the last two PDZ domains, which has been overlooked in the previous studies. Here we characterized a well-folded harmonin homology domain (HHD) from the middle region and solved the PDZD7 HHD structure at the resolution of 1.49 Å. PDZD7 HHD adopts the same five-helix fold as other HHDs found in Harmonin and Whirlin; however, in PDZD7 HHD, a unique α1N helix occupies the canonical binding pocket, suggesting a distinct binding mode. Moreover, we found that the PDZD7 HHD domain can bind lipid and mediate the localization of PDZD7 to the plasma membrane in HEK293T cells. Intriguingly, a hearing-loss mutation at the N-terminal extension region of the HHD can disrupt the lipid-binding ability of PDZD7 HHD, suggesting that HHD-mediated membrane targeting is required for the hearing process. This structural and biochemical characterization of the PDZD7 HHD region provides mechanistic explanations for human deafness-causing mutations in PDZD7. Furthermore, this structure will also facilitate biochemical and functional studies of other HHDs.
Organizational Affiliation:
Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders, Ministry of Education, Bio-X Institutes, Shanghai Jiao Tong University, Shanghai, China.