Common and unique strategies of myoglobin evolution for deep-sea adaptation of diving mammals.
Isogai, Y., Imamura, H., Nakae, S., Sumi, T., Takahashi, K.I., Shirai, T.(2021) iScience 24: 102920-102920
- PubMed: 34430810
- DOI: https://doi.org/10.1016/j.isci.2021.102920
- Primary Citation of Related Structures:
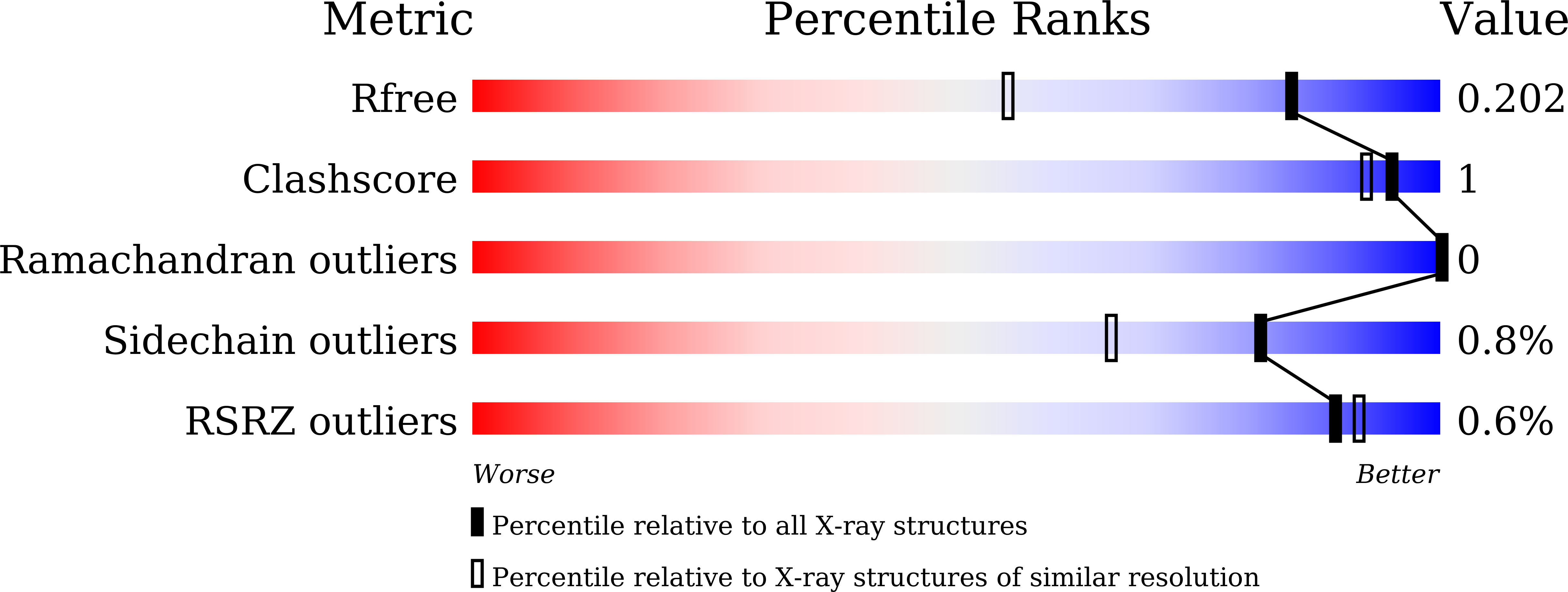
7DDR, 7DDS, 7DDT, 7DDU - PubMed Abstract:
Myoglobin (Mb) is highly concentrated in the myocytes of diving mammals such as whales and seals, in comparison with land animals, and its molecular evolution has played a crucial role in their deep-sea adaptation. We previously resurrected ancestral whale Mbs and demonstrated the evolutional strategies for higher solubility under macromolecular crowding conditions. Pinnipeds, such as seals and sea lions, are also expert diving mammals with Mb-rich muscles. In the present study, we resurrected ancestral pinniped Mbs and investigated their biochemical and structural properties. Comparisons between pinniped and whale Mbs revealed the common and distinctive strategies for the deep-sea adaptation. The overall evolution processes, gaining precipitant tolerance and improving thermodynamic stability, were commonly observed. However, the strategies for improving the folding stability differed, and the pinniped Mbs exploited the shielding of hydrophobic surfaces more effectively than the whale Mbs.
Organizational Affiliation:
Department of Pharmaceutical Engineering, Toyama Prefectural University, Imizu, Toyama 939-0398, Japan.