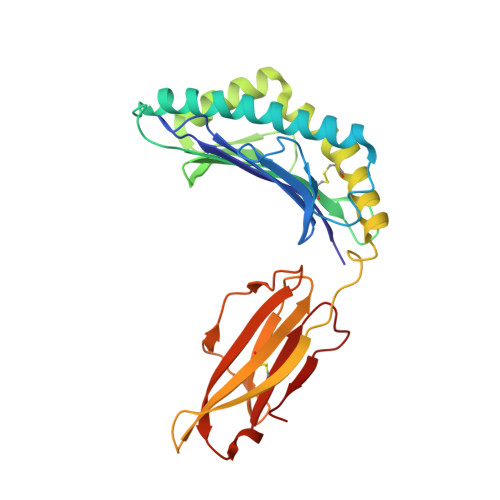
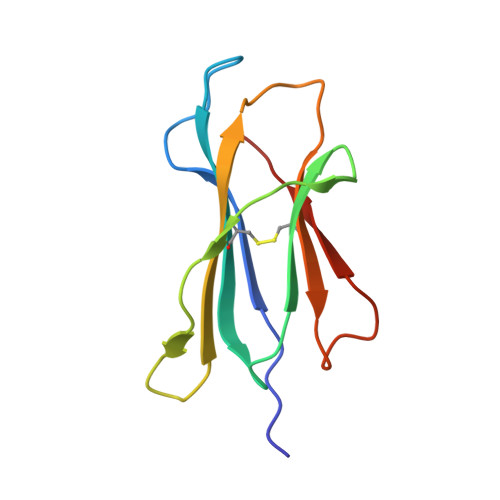
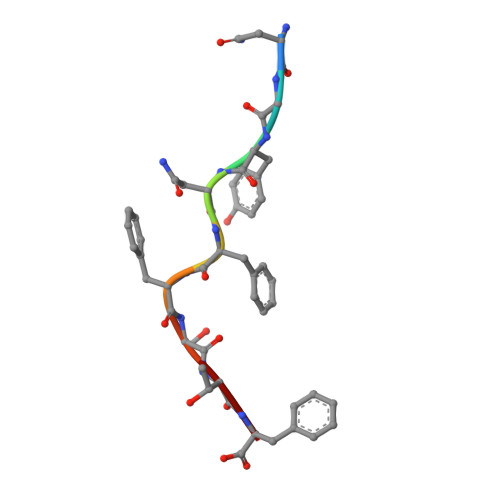
Crystal structure of the giant panda MHC class I complex: First insights into the viral peptide presentation profile in the bear family.
Yuan, H., Ma, L., Zhang, L., Li, X., Xia, C.(2020) Protein Sci 29: 2468-2481
- PubMed: 33078460
- DOI: https://doi.org/10.1002/pro.3980
- Primary Citation of Related Structures:
7DC6 - PubMed Abstract:
The viral cytotoxic T lymphocyte (CTL) epitope peptides presented by classical MHC-I molecules require the assembly of a peptide-MHC-I-β2m (pMHC-I) trimolecular complex for T cell receptor (TCR) recognition, which is the critical activation link for triggering antiviral T cell immunity. Research on T cell immunology in the Ursidae family, especially structural immunology, is still lacking. In this study, the structure of the key trimolecular complex pMHC-I, which binds a peptide from canine distemper virus, was solved for the first time using giant panda as a representative species of Ursidae. The structural characteristics of the giant panda pMHC-I complex (pAime-128), including the unique pockets in the peptide-binding groove (PBG), were analyzed in detail. Comparing the pAime-128 to others in the bear family and extending the comparison to other mammals revealed distinct features. The interaction between MHC-I and β2m, the features of pAime-128 involved in TCR docking and cluster of differentiation 8 (CD8) binding, the anchor sites in the PBG, and the CTL epitopes of potential viruses that infect pandas were clarified. Unique features of pMHC-I viral antigen presentation in the panda were revealed by solving the three-dimensional (3D) structure of pAime-128. The distinct characteristics of pAime-128 indicate an unusual event that emerged during the evolution of the MHC system in the bear family. These results provide a new platform for research on panda CTL immunity and the design of vaccines for application in the bear family.
Organizational Affiliation:
Department of Microbiology and Immunology, College of Veterinary Medicine, China Agricultural University, Beijing, China.