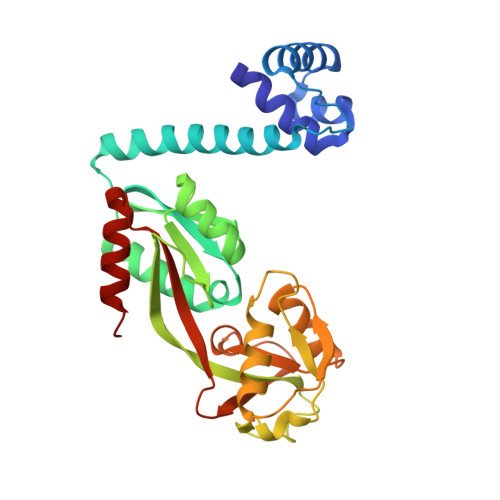
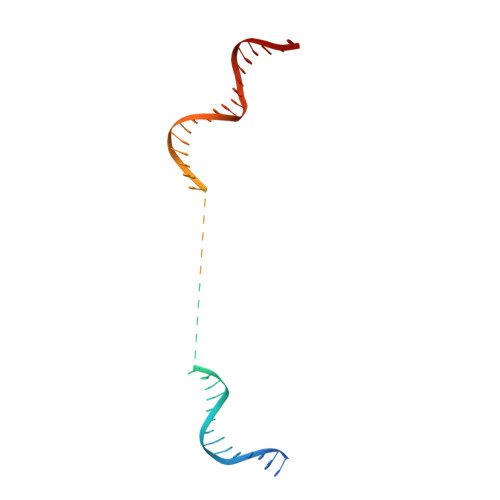
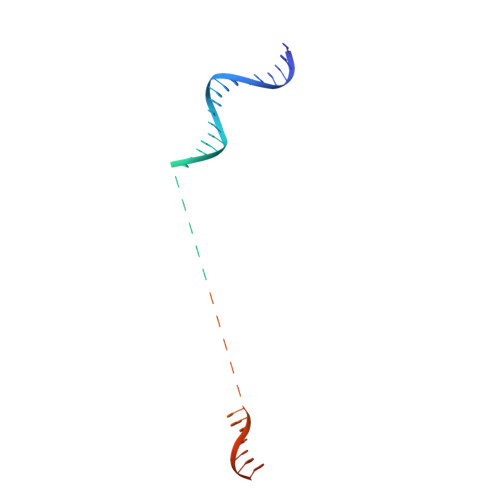
Crystal structure of the full-length LysR-type transcription regulator CbnR in complex with promoter DNA.
Giannopoulou, E.A., Senda, M., Koentjoro, M.P., Adachi, N., Ogawa, N., Senda, T.(2021) FEBS J 288: 4560-4575
- PubMed: 33576566
- DOI: https://doi.org/10.1111/febs.15764
- Primary Citation of Related Structures:
7D98 - PubMed Abstract:
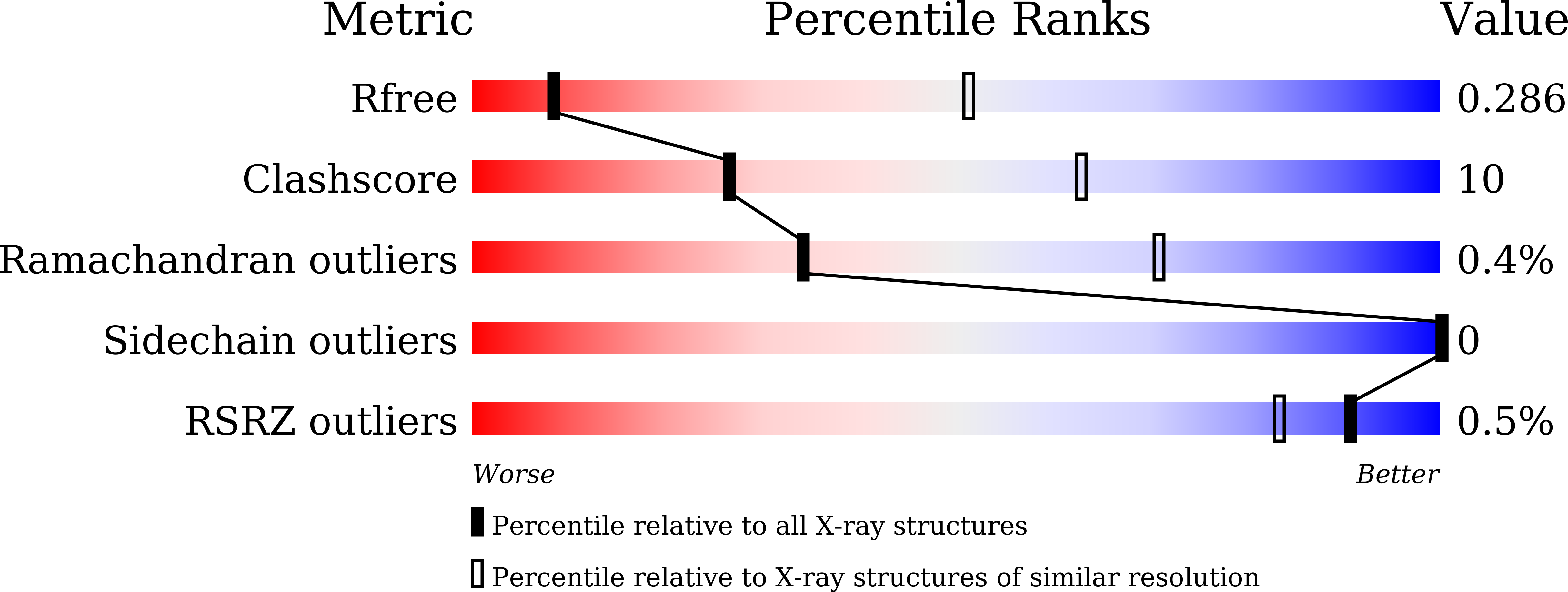
LysR-type transcription regulators (LTTRs) comprise one of the largest families of transcriptional regulators in bacteria. They are typically homo-tetrameric proteins and interact with promoter DNA of ~ 50-60 bp. Earlier biochemical studies have suggested that LTTR binding to promoter DNA bends the DNA and, upon inducer binding, the bend angle of the DNA is reduced through a quaternary structure change of the tetrameric LTTR, leading to the activation of transcription. To date, crystal structures of full-length LTTRs, DNA-binding domains (DBD) with their target DNAs, and the regulatory domains with and without inducer molecules have been reported. However, these crystal structures have not provided direct evidence of the quaternary structure changes of LTTRs or of the molecular mechanism underlying these changes. Here, we report the first crystal structure of a full-length LTTR, CbnR, in complex with its promoter DNA. The crystal structure showed that, in the absence of bound inducer molecules, the four DBDs of the tetrameric CbnR interact with the promoter DNA, bending the DNA by ~ 70°. Structural comparison between the DNA-free and DNA-bound forms demonstrates that the quaternary structure change of the tetrameric CbnR required for promoter region-binding arises from relative orientation changes of the three domains in each subunit. The mechanism of the quaternary structure change caused by inducer binding is also discussed based on the present crystal structure, affinity analysis between CbnR and the promoter DNA, and earlier mutational studies on CbnR. DATABASE: Atomic coordinates and structure factors for the full-length Cupriavidus necator NH9 CbnR in complex with promoter DNA are available in the Protein Data Bank under the accession code 7D98.
Organizational Affiliation:
Structural Biology Research Center, Photon Factory, Institute of Materials Structure Science, High Energy Accelerator Research Organization (KEK), Tsukuba, Japan.