A unique hyperdynamic dimer interface permits small molecule perturbation of the melanoma oncoprotein MITF for melanoma therapy.
Liu, Z., Chen, K., Dai, J., Xu, P., Sun, W., Liu, W., Zhao, Z., Bennett, S.P., Li, P., Ma, T., Lin, Y., Kawakami, A., Yu, J., Wang, F., Wang, C., Li, M., Chase, P., Hodder, P., Spicer, T.P., Scampavia, L., Cao, C., Pan, L., Dong, J., Chen, Y., Yu, B., Guo, M., Fang, P., Fisher, D.E., Wang, J.(2023) Cell Res 33: 55-70
- PubMed: 36588115
- DOI: https://doi.org/10.1038/s41422-022-00744-5
- Primary Citation of Related Structures:
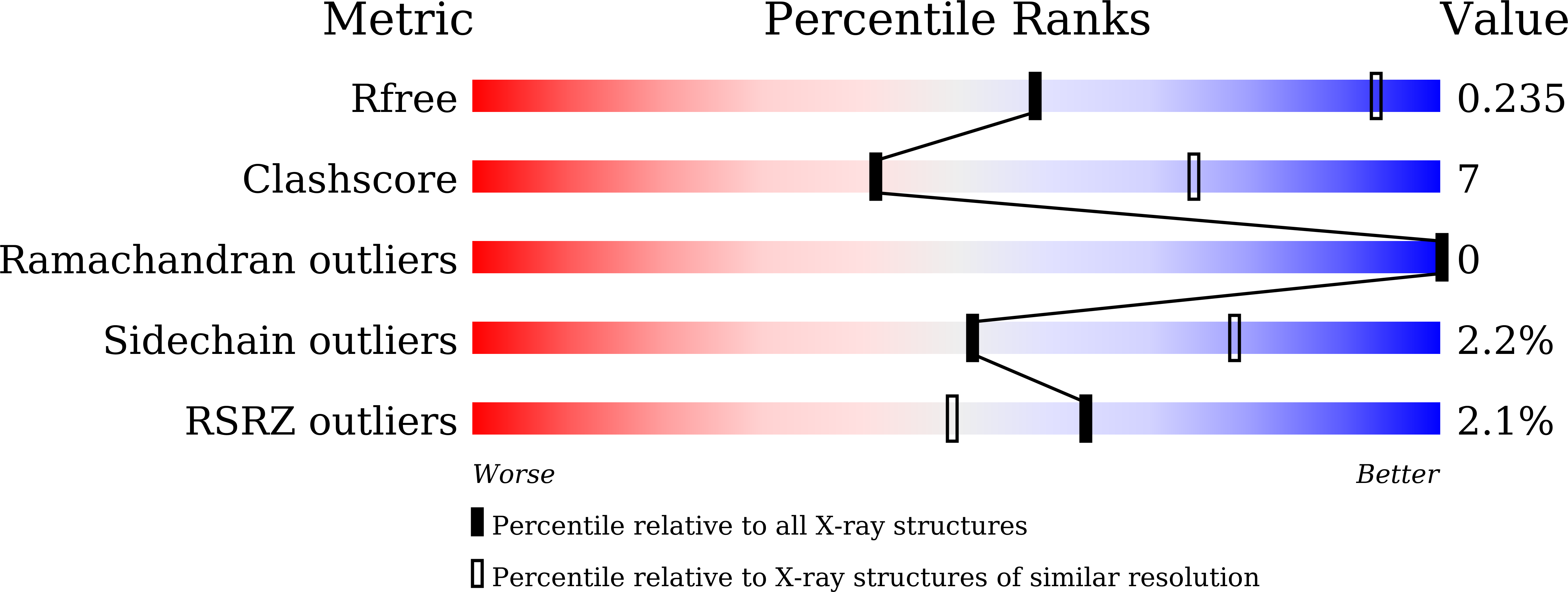
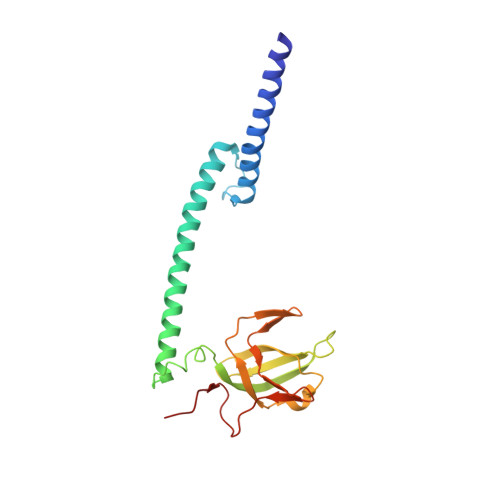
7D8R, 7D8S, 7D8T, 7EOD - PubMed Abstract:
Microphthalmia transcription factor (MITF) regulates melanocyte development and is the "lineage-specific survival" oncogene of melanoma. MITF is essential for melanoma initiation, progression, and relapse and has been considered an important therapeutic target; however, direct inhibition of MITF through small molecules is considered impossible, due to the absence of a ligand-binding pocket for drug design. Here, our structural analyses show that the structure of MITF is hyperdynamic because of its out-of-register leucine zipper with a 3-residue insertion. The dynamic MITF is highly vulnerable to dimer-disrupting mutations, as we observed that MITF loss-of-function mutations in human Waardenburg syndrome type 2 A are frequently located on the dimer interface and disrupt the dimer forming ability accordingly. These observations suggest a unique opportunity to inhibit MITF with small molecules capable of disrupting the MITF dimer. From a high throughput screening against 654,650 compounds, we discovered compound TT-012, which specifically binds to dynamic MITF and destroys the latter's dimer formation and DNA-binding ability. Using chromatin immunoprecipitation assay and RNA sequencing, we showed that TT-012 inhibits the transcriptional activity of MITF in B16F10 melanoma cells. In addition, TT-012 inhibits the growth of high-MITF melanoma cells, and inhibits the tumor growth and metastasis with tolerable toxicity to liver and immune cells in animal models. Together, this study demonstrates a unique hyperdynamic dimer interface in melanoma oncoprotein MITF, and reveals a novel approach to therapeutically suppress MITF activity.
Organizational Affiliation:
State Key Laboratory of Bioorganic and Natural Products Chemistry, Center for Excellence in Molecular Synthesis, Shanghai Institute of Organic Chemistry, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai, China.