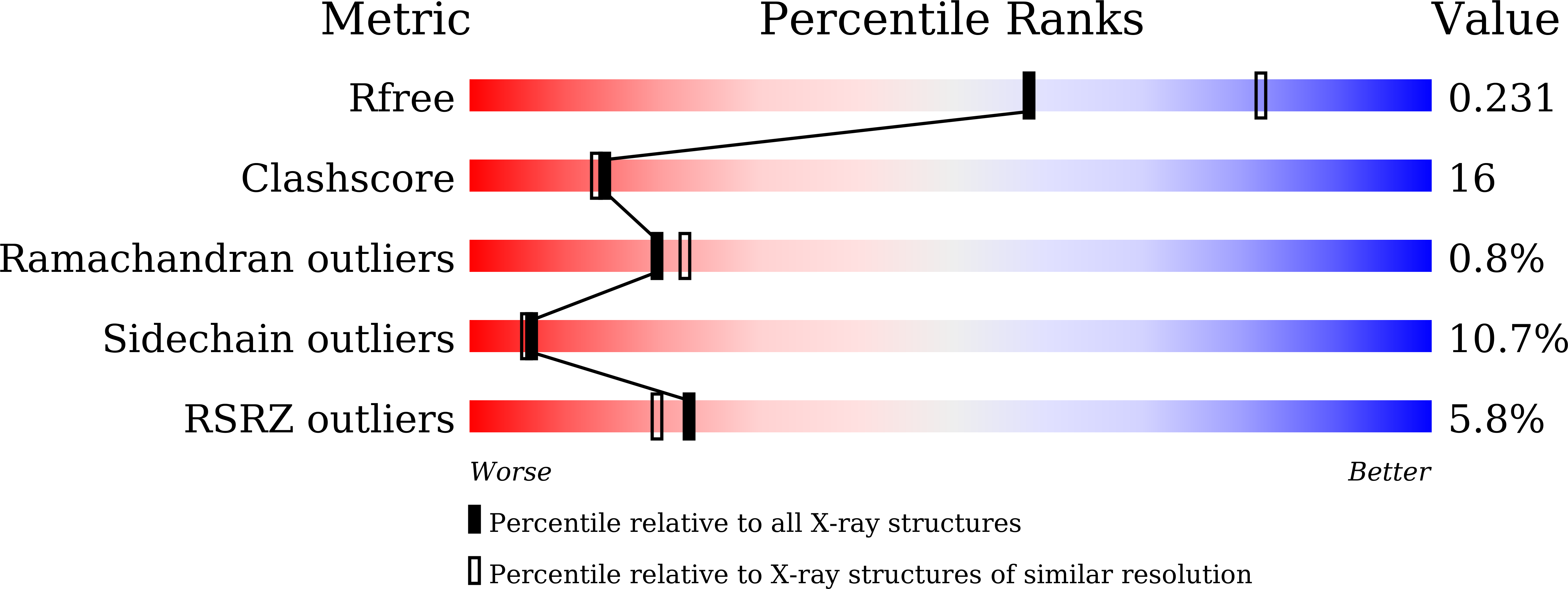
Discovery of novel HIV-1 integrase-LEDGF/p75 allosteric inhibitors based on a pyridine scaffold forming an intramolecular hydrogen bond.
Sugiyama, S., Akiyama, T., Taoda, Y., Iwaki, T., Matsuoka, E., Akihisa, E., Seki, T., Yoshinaga, T., Kawasuji, T.(2020) Bioorg Med Chem Lett 33: 127742-127742
- PubMed: 33316407
- DOI: https://doi.org/10.1016/j.bmcl.2020.127742
- Primary Citation of Related Structures:
7D83 - PubMed Abstract:
We have discovered HIV-1 novel integrase-LEDGF/p75 allosteric inhibitors (INLAIs) based on a pyridine scaffold forming an intramolecular hydrogen bond. Scaffolds containing a pyridine moiety have been studied extensively and we have already reported that substituents extending from the C1 position contributed to the antiviral potency. In this study, we designed a new pyridine scaffold 2 with a substituent at the C1 position. Interestingly, during attempts at optimization, we found that the direction of the C1 substituents with an intramolecular hydrogen bond contributed to the antiviral potency. Compound 34f exhibited better antiviral potency against WT and the T174I mutant (EC 50 (WT) = 6.6 nM, EC 50 (T174I) = 270 nM) than BI 224436 (EC 50 (WT) = 22 nM, EC 50 (T174I) > 5000 nM).
Organizational Affiliation:
Shionogi Pharmaceutical Research Center, Shionogi & Co., Ltd., 3-1-1, Futabacho, Toyonaka, Osaka 561-0825, Japan. Electronic address: shuichi.sugiyama@shionogi.co.jp.