A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor.
Sun, W., Chen, L.N., Zhou, Q., Zhao, L.H., Yang, D., Zhang, H., Cong, Z., Shen, D.D., Zhao, F., Zhou, F., Cai, X., Chen, Y., Zhou, Y., Gadgaard, S., van der Velden, W.J.C., Zhao, S., Jiang, Y., Rosenkilde, M.M., Xu, H.E., Zhang, Y., Wang, M.W.(2020) Cell Res 30: 1098-1108
- PubMed: 33239759
- DOI: https://doi.org/10.1038/s41422-020-00442-0
- Primary Citation of Related Structures:
7D68 - PubMed Abstract:
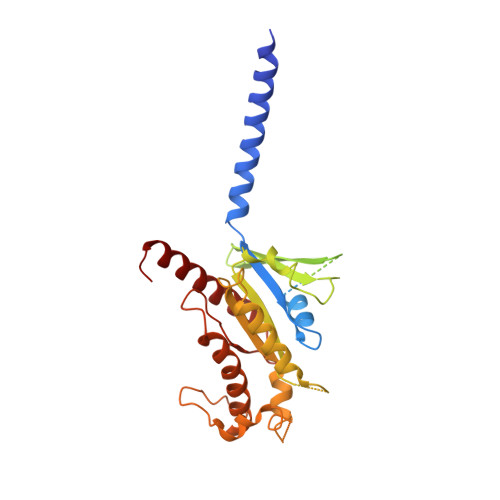
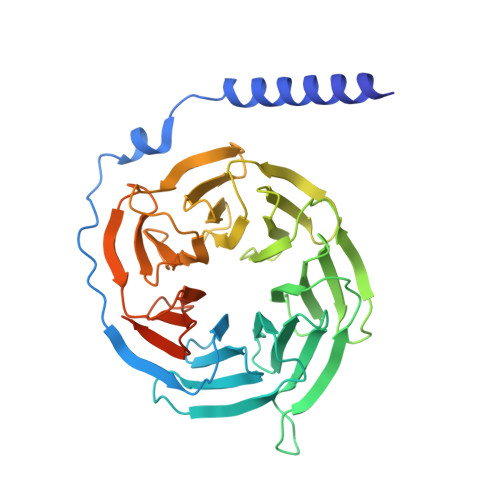
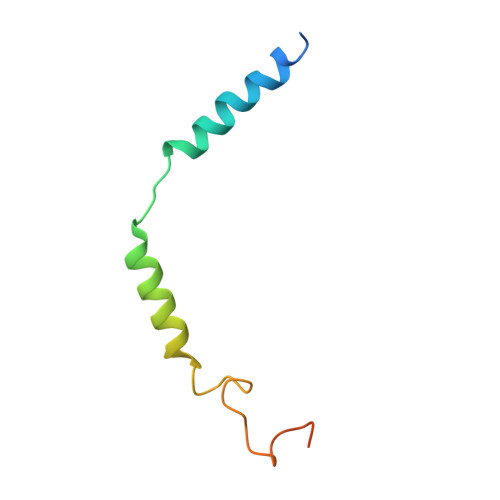
Glucagon-like peptides (GLP-1 and GLP-2) are two proglucagon-derived intestinal hormones that mediate distinct physiological functions through two related receptors (GLP-1R and GLP-2R) which are important drug targets for metabolic disorders and Crohn's disease, respectively. Despite great progress in GLP-1R structure determination, our understanding on the differences of peptide binding and signal transduction between these two receptors remains elusive. Here we report the electron microscopy structure of the human GLP-2R in complex with GLP-2 and a G s heterotrimer. To accommodate GLP-2 rather than GLP-1, GLP-2R fine-tunes the conformations of the extracellular parts of transmembrane helices (TMs) 1, 5, 7 and extracellular loop 1 (ECL1). In contrast to GLP-1, the N-terminal histidine of GLP-2 penetrates into the receptor core with a unique orientation. The middle region of GLP-2 engages with TM1 and TM7 more extensively than with ECL2, and the GLP-2 C-terminus closely attaches to ECL1, which is the most protruded among 9 class B G protein-coupled receptors (GPCRs). Functional studies revealed that the above three segments of GLP-2 are essential for GLP-2 recognition and receptor activation, especially the middle region. These results provide new insights into the molecular basis of ligand specificity in class B GPCRs and may facilitate the development of more specific therapeutics.
Organizational Affiliation:
The CAS Key Laboratory of Receptor Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, 201203, China.