Cryo-EM structure of mycobacterial cytochrome bd reveals two oxygen access channels.
Wang, W., Gao, Y., Tang, Y., Zhou, X., Lai, Y., Zhou, S., Zhang, Y., Yang, X., Liu, F., Guddat, L.W., Wang, Q., Rao, Z., Gong, H.(2021) Nat Commun 12: 4621-4621
- PubMed: 34330928
- DOI: https://doi.org/10.1038/s41467-021-24924-w
- Primary Citation of Related Structures:
7D5I - PubMed Abstract:
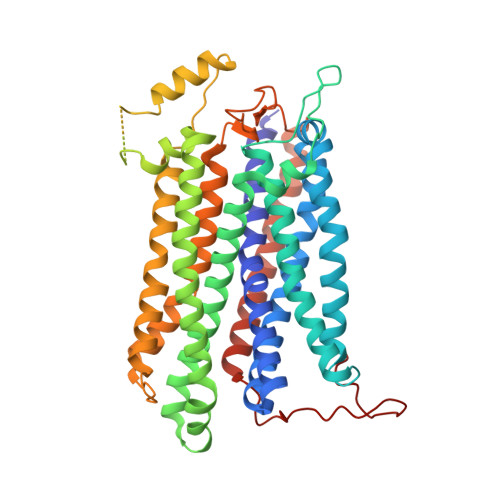
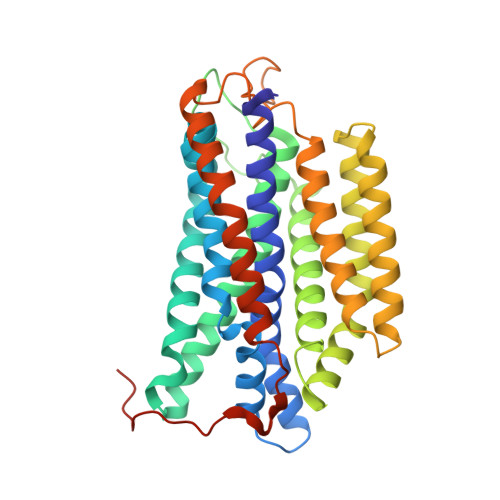
Cytochromes bd are ubiquitous amongst prokaryotes including many human-pathogenic bacteria. Such complexes are targets for the development of antimicrobial drugs. However, an understanding of the relationship between the structure and functional mechanisms of these oxidases is incomplete. Here, we have determined the 2.8 Å structure of Mycobacterium smegmatis cytochrome bd by single-particle cryo-electron microscopy. This bd oxidase consists of two subunits CydA and CydB, that adopt a pseudo two-fold symmetrical arrangement. The structural topology of its Q-loop domain, whose function is to bind the substrate, quinol, is significantly different compared to the C-terminal region reported for cytochromes bd from Geobacillus thermodenitrificans (G. th) and Escherichia coli (E. coli). In addition, we have identified two potential oxygen access channels in the structure and shown that similar tunnels also exist in G. th and E. coli cytochromes bd. This study provides insights to develop a framework for the rational design of antituberculosis compounds that block the oxygen access channels of this oxidase.
Organizational Affiliation:
Shanghai Institute for Advanced Immunochemical Studies and School of Life Science and Technology, ShanghaiTech University, Shanghai, China.