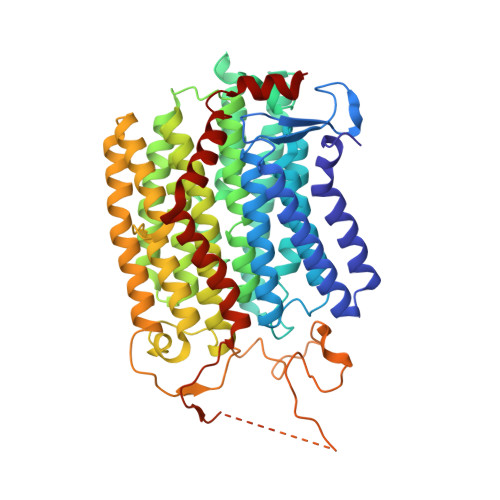
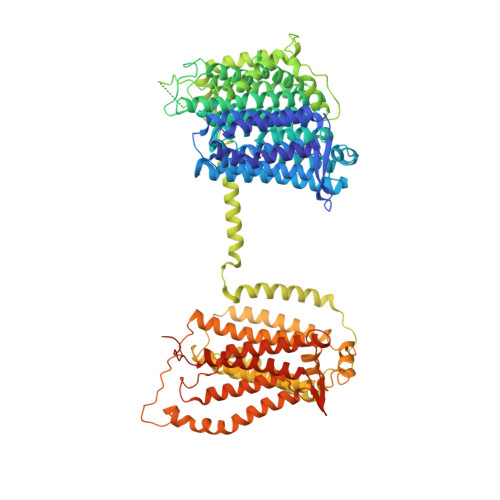
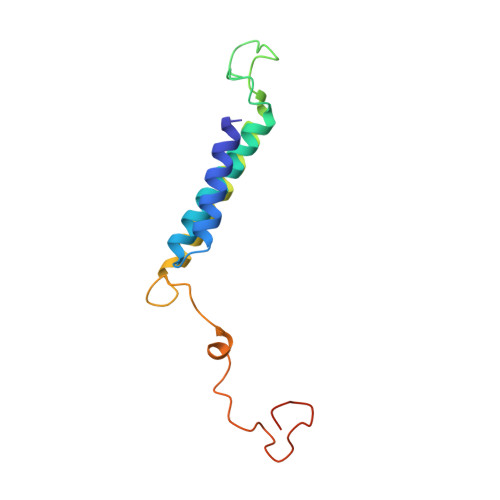
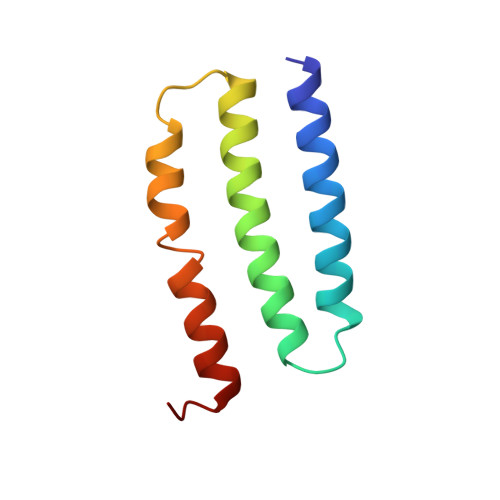
Structure of the Dietzia Mrp complex reveals molecular mechanism of this giant bacterial sodium proton pump.
Li, B., Zhang, K., Nie, Y., Wang, X., Zhao, Y., Zhang, X.C., Wu, X.L.(2020) Proc Natl Acad Sci U S A 117: 31166-31176
- PubMed: 33229520
- DOI: https://doi.org/10.1073/pnas.2006276117
- Primary Citation of Related Structures:
7D3U - PubMed Abstract:
Multiple resistance and pH adaptation (Mrp) complexes are sophisticated cation/proton exchangers found in a vast variety of alkaliphilic and/or halophilic microorganisms, and are critical for their survival in highly challenging environments. This family of antiporters is likely to represent the ancestor of cation pumps found in many redox-driven transporter complexes, including the complex I of the respiratory chain. Here, we present the three-dimensional structure of the Mrp complex from a Dietzia sp. strain solved at 3.0-Å resolution using the single-particle cryoelectron microscopy method. Our structure-based mutagenesis and functional analyses suggest that the substrate translocation pathways for the driving substance protons and the substrate sodium ions are separated in two modules and that symmetry-restrained conformational change underlies the functional cycle of the transporter. Our findings shed light on mechanisms of redox-driven primary active transporters, and explain how driving substances of different electric charges may drive similar transport processes.
Organizational Affiliation:
National Laboratory of Biomacromolecules, Chinese Academy of Sciences Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, 100101 Beijing, China.