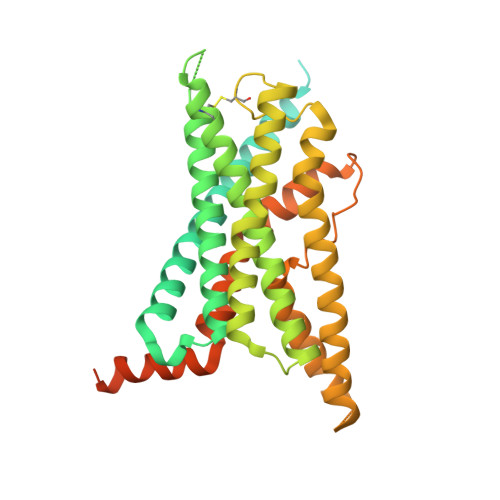
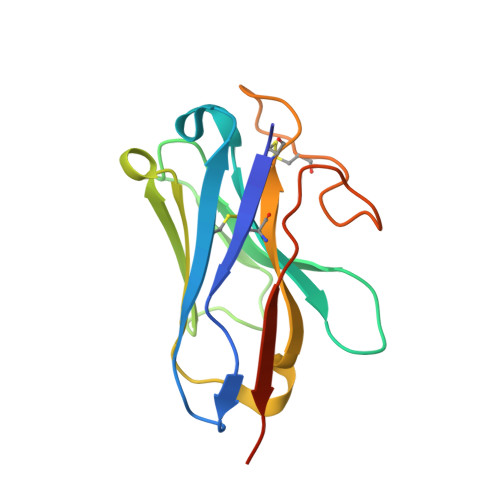
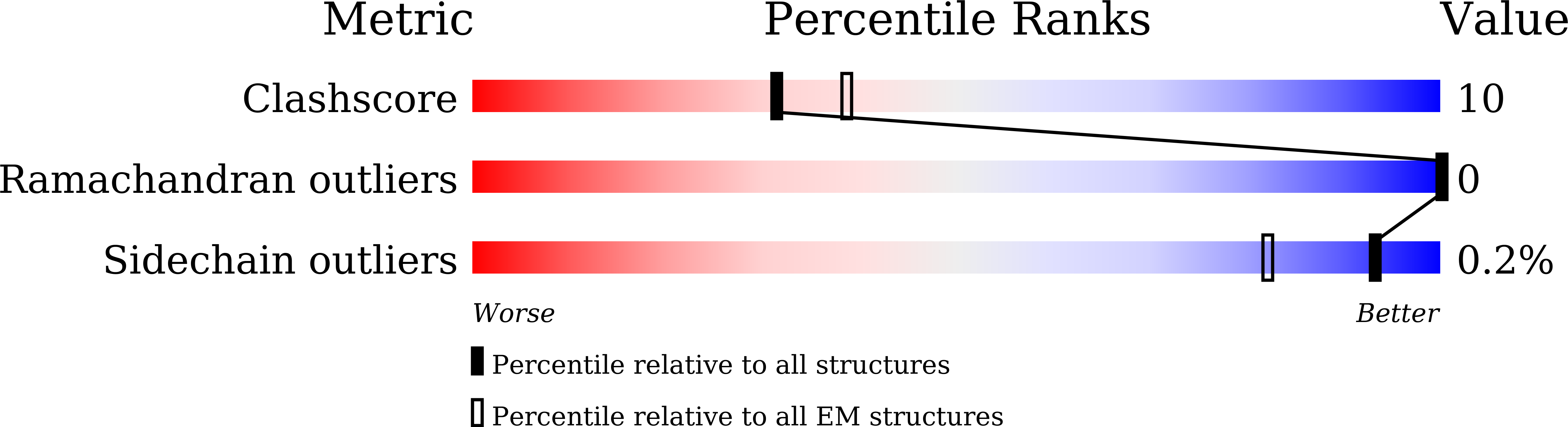Structure of the human secretin receptor coupled to an engineered heterotrimeric G protein.
Fukuhara, S., Kobayashi, K., Kusakizako, T., Iida, W., Kato, M., Shihoya, W., Nureki, O.(2020) Biochem Biophys Res Commun 533: 861-866
- PubMed: 33008599
- DOI: https://doi.org/10.1016/j.bbrc.2020.08.042
- Primary Citation of Related Structures:
7D3S - PubMed Abstract:
Secretin is a gastrointestinal hormone that exerts multiple physiological functions via activation of the secretin receptor (SECR). SECR belongs to the class B G-protein-coupled receptors and is involved in various processes, such as regulation of the pH of the duodenal content, food intake, and water homeostasis. Here, we report a cryo-electron microscopy structure of human SECR bound to secretin and an engineered Gs heterotrimer. The structure revealed the basic architecture of SECR and the secretin binding mode. A structural comparison of the SECR and PAC1R transmembrane domains revealed that transmembrane helices 1 and 2 play a prominent role in secretin recognition. Moreover, the extracellular domain of SECR is perpendicular to the TMD, unlike that of PAC1R. This comparison revealed the diverged peptide recognition mechanisms of these receptors, which belong to the same subgroup. Our structural information will facilitate drug discovery research for clinical applications.
Organizational Affiliation:
Department of Biological Sciences, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, 113-0033, Japan; Department of Family Medicine, Graduate School of Medical and Dental Sciences, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo, 113-8519, Japan.