An improved fluorescent protein tag and its nanobodies for membrane protein expression, stability assay, and purification
Cai, H., Yao, H., Li, T., Hutter, C., Tang, Y., Li, Y., Seeger, M., Li, D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Thermostable green fluorescent protein (TGP) | 232 | Galaxea fascicularis | Mutation(s): 0 | 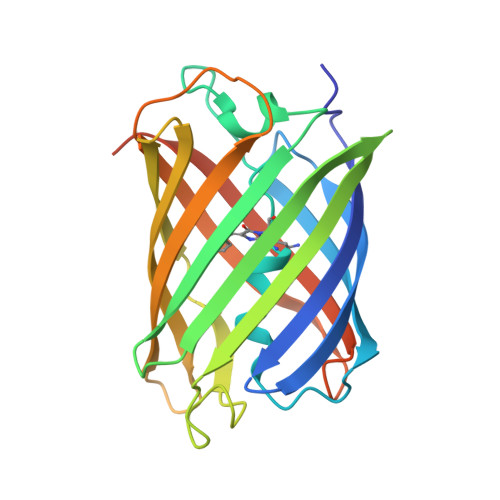 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Synthetic nanobody (Sybody) 92 recognizing the thermostable green fluorescent protein (TGP) | 144 | synthetic construct | Mutation(s): 0 | 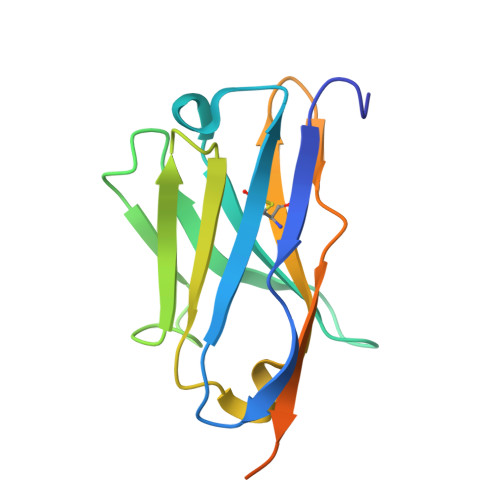 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CAD Query on CAD | L [auth A] | CACODYLIC ACID C2 H7 As O2 OGGXGZAMXPVRFZ-UHFFFAOYSA-N |  | ||
| CAC Query on CAC | U [auth D] | CACODYLATE ION C2 H6 As O2 OGGXGZAMXPVRFZ-UHFFFAOYSA-M |  | ||
| GOL Query on GOL | I [auth A] J [auth A] K [auth A] M [auth A] N [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | P [auth B], W [auth D] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CRQ Query on CRQ | A, B, C, D | L-PEPTIDE LINKING | C16 H16 N4 O5 |  | GLN, TYR, GLY |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 95.245 | α = 90 |
| b = 130.154 | β = 90 |
| c = 171.453 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Science Foundation (NSF, China) | China | 31870726 |