Structural basis for the recognition of human hemoglobin by the Shr protein from Streptococcus pyogenes
Caaveiro, J.M.M., Hoshino, M., Senoo, A., Nakakido, M., Nagatoishi, S., Tame, J.R.H., Tsumoto, K.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hemoglobin subunit alpha | 142 | Homo sapiens | Mutation(s): 0 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P69905 (Homo sapiens) Explore P69905 Go to UniProtKB: P69905 | |||||
PHAROS: P69905 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P69905 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hemoglobin subunit beta | 147 | Homo sapiens | Mutation(s): 0 |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P68871 (Homo sapiens) Explore P68871 Go to UniProtKB: P68871 | |||||
PHAROS: P68871 GTEx: ENSG00000244734 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68871 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Amino acid ABC transporter substrate-binding protein | E, F, G [auth H] | 124 | Streptococcus pyogenes | Mutation(s): 0 Gene Names: shr, inlA_2, G3M98_06270, GTK50_07335, SAMEA3919037_01262 | 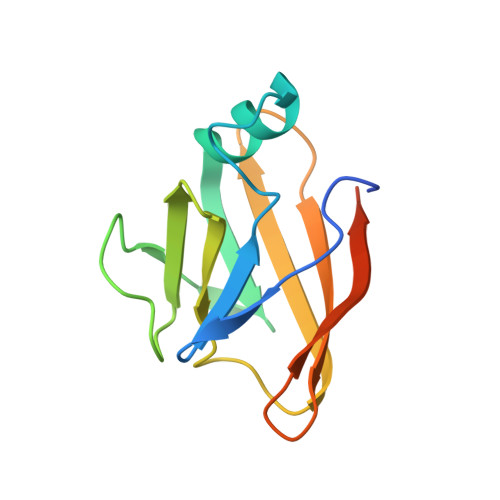 |
UniProt | |||||
Find proteins for B0LFQ8 (Streptococcus pyogenes) Explore B0LFQ8 Go to UniProtKB: B0LFQ8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B0LFQ8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEM Query on HEM | H [auth A], I [auth B], K [auth C], L [auth D] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| SO4 Query on SO4 | J [auth B], M [auth F] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 178.5 | α = 90 |
| b = 52.76 | β = 118.35 |
| c = 129.17 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| MOSFLM | data reduction |
| SCALA | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | 15K06962 |