Moving toward generalizable NZ-1 labeling for 3D structure determination with optimized epitope-tag insertion.
Tamura-Sakaguchi, R., Aruga, R., Hirose, M., Ekimoto, T., Miyake, T., Hizukuri, Y., Oi, R., Kaneko, M.K., Kato, Y., Akiyama, Y., Ikeguchi, M., Iwasaki, K., Nogi, T.(2021) Acta Crystallogr D Struct Biol 77: 645-662
- PubMed: 33950020
- DOI: https://doi.org/10.1107/S2059798321002527
- Primary Citation of Related Structures:
7CQC, 7CQD - PubMed Abstract:
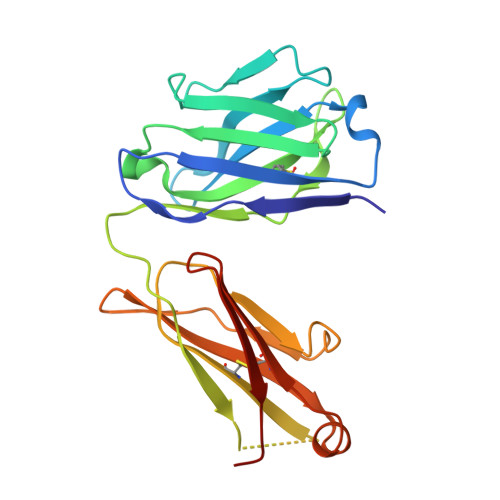
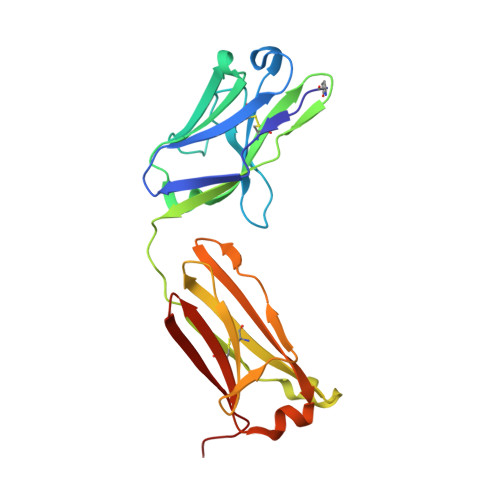
Antibody labeling has been conducted extensively for structure determination using both X-ray crystallography and electron microscopy (EM). However, establishing target-specific antibodies is a prerequisite for applying antibody-assisted structural analysis. To expand the applicability of this strategy, an alternative method has been developed to prepare an antibody complex by inserting an exogenous epitope into the target. It has already been demonstrated that the Fab of the NZ-1 monoclonal antibody can form a stable complex with a target containing a PA12 tag as an inserted epitope. Nevertheless, it was also found that complex formation through the inserted PA12 tag inevitably caused structural changes around the insertion site on the target. Here, an attempt was made to improve the tag-insertion method, and it was consequently discovered that an alternate tag (PA14) could replace various loops on the target without inducing large structural changes. Crystallographic analysis demonstrated that the inserted PA14 tag adopts a loop-like conformation with closed ends in the antigen-binding pocket of the NZ-1 Fab. Due to proximity of the termini in the bound conformation, the more optimal PA14 tag had only a minor impact on the target structure. In fact, the PA14 tag could also be inserted into a sterically hindered loop for labeling. Molecular-dynamics simulations also showed a rigid structure for the target regardless of PA14 insertion and complex formation with the NZ-1 Fab. Using this improved labeling technique, negative-stain EM was performed on a bacterial site-2 protease, which enabled an approximation of the domain arrangement based on the docking mode of the NZ-1 Fab.
Organizational Affiliation:
Graduate School of Medical Life Science, Yokohama City University, Japan.