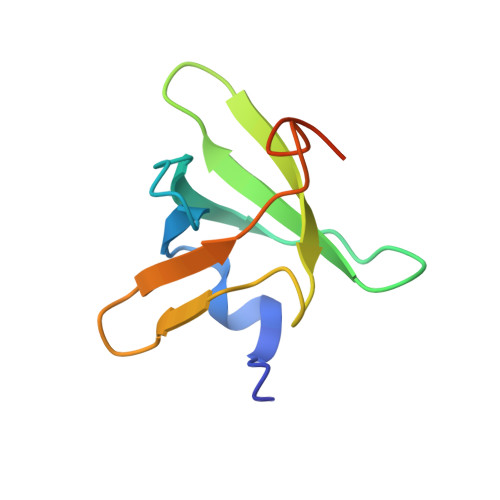
Structural Basis for the C-Terminal Domain of Mycobacterium tuberculosis Ribosome Maturation Factor RimM to Bind Ribosomal Protein S19.
Zhang, H., Zhou, Q., Guo, C., Feng, L., Wang, H., Liao, X., Lin, D.(2021) Biomolecules 11
- PubMed: 33919647
- DOI: https://doi.org/10.3390/biom11040597
- Primary Citation of Related Structures:
7CQ1 - PubMed Abstract:
Multidrug-resistant tuberculosis (TB) is a serious threat to public health, calling for the development of new anti-TB drugs. Chaperon protein RimM, involved in the assembly of ribosomal protein S19 into 30S ribosomal subunit during ribosome maturation, is a potential drug target for TB treatment. The C-terminal domain (CTD) of RimM is primarily responsible for binding S19. However, both the CTD structure of RimM from Mycobacterium tuberculosis ( Mtb RimM CTD ) and the molecular mechanisms underlying Mtb RimM CTD binding S19 remain elusive. Here, we report the solution structure, dynamics features of Mtb RimM CTD , and its interaction with S19. Mtb RimM CTD has a rigid hydrophobic core comprised of a relatively conservative six-strand β-barrel, tailed with a short α-helix and interspersed with flexible loops. Using several biophysical techniques including surface plasmon resonance (SPR) affinity assays, nuclear magnetic resonance (NMR) assays, and molecular docking, we established a structural model of the Mtb RimM CTD -S19 complex and indicated that the β4-β5 loop and two nonconserved key residues (D105 and H129) significantly contributed to the unique pattern of Mtb RimM CTD binding S19, which might be implicated in a form of orthogonality for species-dependent RimM-S19 interaction. Our study provides the structural basis for Mtb RimM CTD binding S19 and is beneficial to the further exploration of Mtb RimM as a potential target for the development of new anti-TB drugs.
Organizational Affiliation:
MOE Key Laboratory of Spectrochemical Analysis & Instrumentation, Key Laboratory of Chemical Biology of Fujian Province, College of Chemistry and Chemical Engineering, Xiamen University, Xiamen 361005, China.