De Novo Design of Allosteric Control into Rotary Motor V1-ATPase by Restoring Lost Function
Kosugi, T., Iida, T., Tanabe, M., Iino, R., Koga, N.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type sodium ATPase catalytic subunit A | 596 | Enterococcus hirae ATCC 9790 | Mutation(s): 0 Gene Names: ntpA, EHR_08260 EC: 7.2.2.1 | 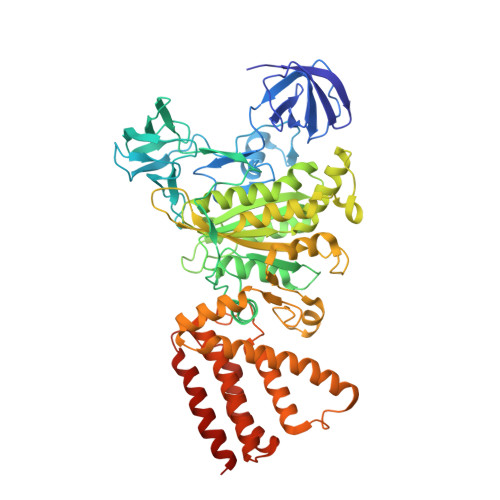 | |
UniProt | |||||
Find proteins for Q08636 (Enterococcus hirae (strain ATCC 9790 / DSM 20160 / JCM 8729 / LMG 6399 / NBRC 3181 / NCIMB 6459 / NCDO 1258 / NCTC 12367 / WDCM 00089 / R)) Explore Q08636 Go to UniProtKB: Q08636 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q08636 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| V-type sodium ATPase subunit B | 458 | Enterococcus hirae ATCC 9790 | Mutation(s): 10 Gene Names: ntpB, EHR_08265 | 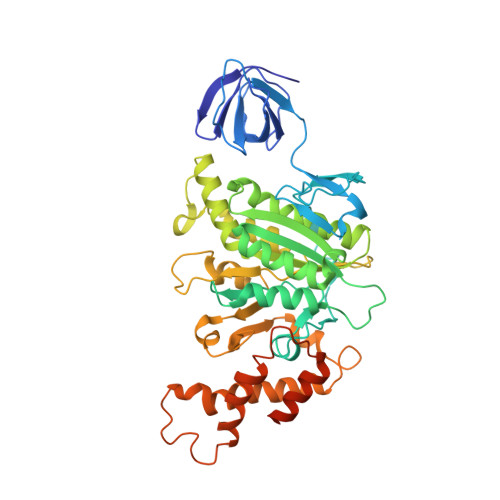 | |
UniProt | |||||
Find proteins for Q08637 (Enterococcus hirae (strain ATCC 9790 / DSM 20160 / JCM 8729 / LMG 6399 / NBRC 3181 / NCIMB 6459 / NCDO 1258 / NCTC 12367 / WDCM 00089 / R)) Explore Q08637 Go to UniProtKB: Q08637 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q08637 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ANP (Subject of Investigation/LOI) Query on ANP | H [auth C] | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER C10 H17 N6 O12 P3 PVKSNHVPLWYQGJ-KQYNXXCUSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | G [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 124.445 | α = 90 |
| b = 124.486 | β = 93.03 |
| c = 131.339 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data processing |
| Aimless | data scaling |
| PHASER | phasing |
| XDS | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | 18H05420 |
| Japan Agency for Medical Research and Development (AMED) | Japan | JP20am0101071 |
| Japan Science and Technology | Japan | JPMJPR13AD |