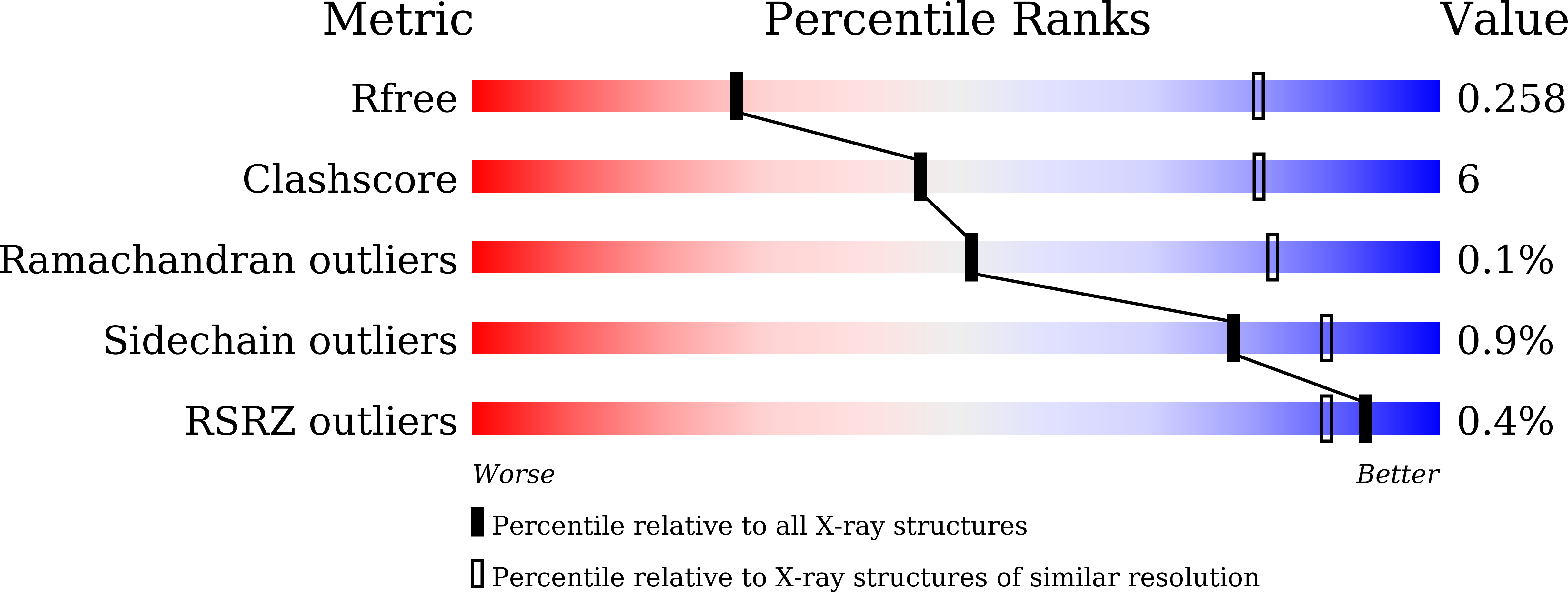
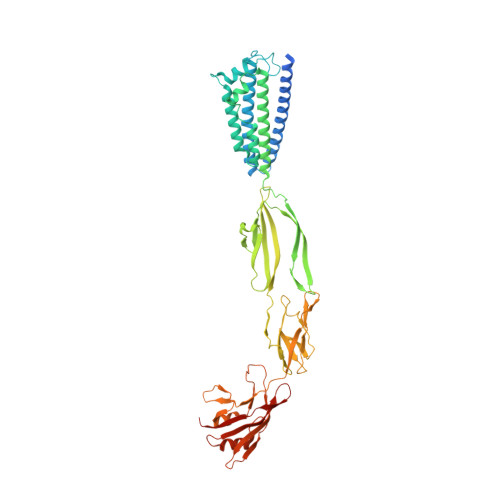
PorM, a core component of bacterial type IX secretion system, forms a dimer with a unique kinked-rod shape.
Sato, K., Okada, K., Nakayama, K., Imada, K.(2020) Biochem Biophys Res Commun 532: 114-119
- PubMed: 32828535
- DOI: https://doi.org/10.1016/j.bbrc.2020.08.018
- Primary Citation of Related Structures:
7CMG - PubMed Abstract:
Porphyromonas gingivalis, which is a major pathogen of the periodontal disease, secrets virulence factors such as gingipain proteases via the type IX secretion system (T9SS). T9SS consists of a trans-periplasmic core complex, the outer membrane translocon complex and the cell-surface complex attached on the outer membrane. PorM is a major component of the trans-periplasmic core complex and is believed to connect the outer membrane component with the inner membrane component. Recent structural studies have revealed that the periplasmic region of GldM, a PorM homolog of a gliding bacterium, consist of four domains and forms a dimer with a straight rod shape. However, only fragment structures are known for PorM. Moreover, one of the PorM fragment structure shows a kink. Here we show the structure of the entire structure of the periplasmic region of PorM (PorMp) at 3.7 Å resolution. PorMp is made up of four domains and forms a unique dimeric structure with an asymmetric, kinked-rod shape. The structure and the following mutational analysis revealed that R204 stabilizes the kink between the D1 and D2 domains and is essential for gingipains secretion, suggesting that the kinked structure of PorM is important for the functional T9SS formation.
Organizational Affiliation:
Department of Microbiology and Oral Infection, Graduate School of Biomedical Sciences, Nagasaki University, Nagasaki, Japan.