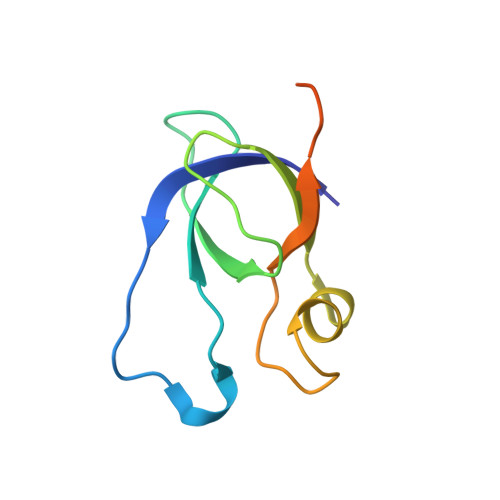
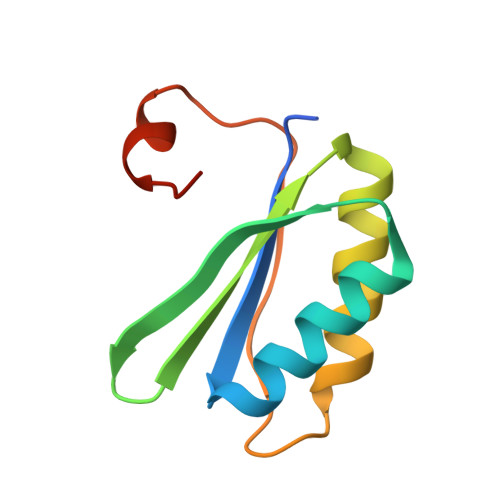
Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Tan, Y.Q., Ali, S., Xue, B., Teo, W.Z., Ling, L.H., Go, M.K., Lv, H., Robinson, R.C., Narita, A., Yew, W.S.(2021) Biomacromolecules 22: 4095-4109
- PubMed: 34384019
- DOI: https://doi.org/10.1021/acs.biomac.1c00533
- Primary Citation of Related Structures:
7CKB, 7CKC, 7DHQ - PubMed Abstract:
Bacterial microcompartments are proteinaceous shells that encase specialized metabolic processes in bacteria. Recent advances in simplification of these intricate shells have encouraged bioengineering efforts. Here, we construct minimal shells derived from the Halothiobacillus neapolitanus α-carboxysome, which we term Cso-shell. Using cryogenic electron microscopy, the atomic-level structures of two shell forms were obtained, reinforcing notions of evolutionarily conserved features in bacterial microcompartment shell architecture. Encapsulation peptide sequences that facilitate loading of heterologous protein cargo within the shells were identified. We further provide a first demonstration in utilizing minimal bacterial microcompartment-derived shells for hosting heterologous enzymes. Cso-shells were found to stabilize enzymatic activities against heat shock, presence of methanol co-solvent, consecutive freeze-thawing, and alkaline environments. This study yields insights into α-carboxysome assembly and advances the utility of synthetic bacterial microcompartments as nanoreactors capable of stabilizing enzymes with varied properties and reaction chemistries.
Organizational Affiliation:
Department of Biochemistry, Yong Loo Lin School of Medicine, National University of Singapore (NUS), 8 Medical Drive, Singapore 117597.