DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Yu, H., Li, J., Liu, G., Zhao, G., Wang, Y., Hu, W., Deng, Z., Wu, G., Gan, J., Zhao, Y.L., He, X.(2020) Nucleic Acids Res 48: 8755-8766
- PubMed: 32621606
- DOI: https://doi.org/10.1093/nar/gkaa574
- Primary Citation of Related Structures:
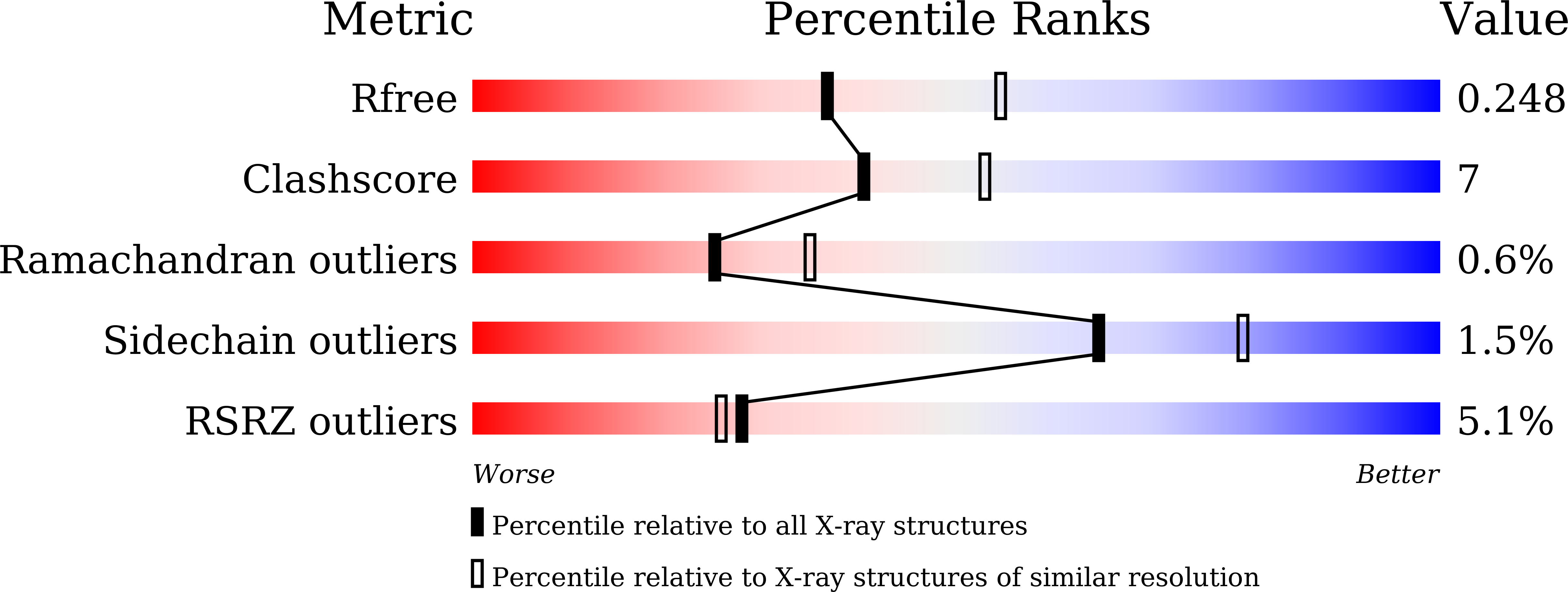
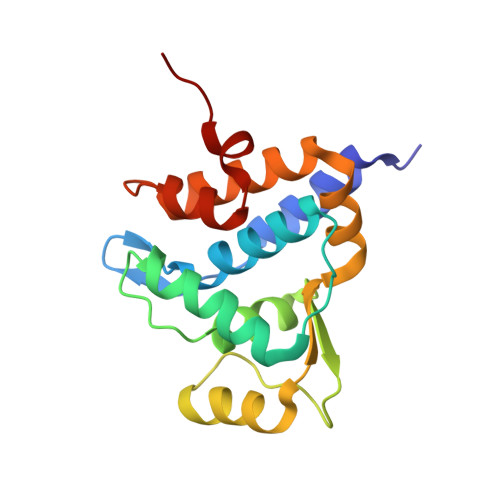
7CC9, 7CCD, 7CCJ - PubMed Abstract:
The sulfur atom of phosphorothioated DNA (PT-DNA) is coordinated by a surface cavity in the conserved sulfur-binding domain (SBD) of type IV restriction enzymes. However, some SBDs cannot recognize the sulfur atom in some sequence contexts. To illustrate the structural determinants for sequence specificity, we resolved the structure of SBDSpr, from endonuclease SprMcrA, in complex with DNA of GPSGCC, GPSATC and GPSAAC contexts. Structural and computational analyses explained why it binds the above PT-DNAs with an affinity in a decreasing order. The structural analysis of SBDSpr-GPSGCC and SBDSco-GPSGCC, the latter only recognizes DNA of GPSGCC, revealed that a positively charged loop above the sulfur-coordination cavity electrostatically interacts with the neighboring DNA phosphate linkage. The structural analysis indicated that the DNA-protein hydrogen bonding pattern and weak non-bonded interaction played important roles in sequence specificity of SBD protein. Exchanges of the positively-charged amino acid residues with the negatively-charged residues in the loop would enable SBDSco to extend recognization for more PT-DNA sequences, implying that type IV endonucleases can be engineered to recognize PT-DNA in novel target sequences.
Organizational Affiliation:
State Key Laboratory of Microbial Metabolism, Joint International Research Laboratory of Metabolic & Developmental Sciences, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, 800 Dongchuan Road, Shanghai 200240, People's Republic of China.