Structural and biochemical analyses of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila involved in mucin degradation.
Xu, W., Yang, W., Wang, Y., Wang, M., Zhang, M.(2020) Biochem Biophys Res Commun 529: 876-881
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-N-acetylhexosaminidase | 538 | Akkermansia muciniphila ATCC BAA-835 | Mutation(s): 0 Gene Names: Amuc_0868 EC: 3.2.1.52 | 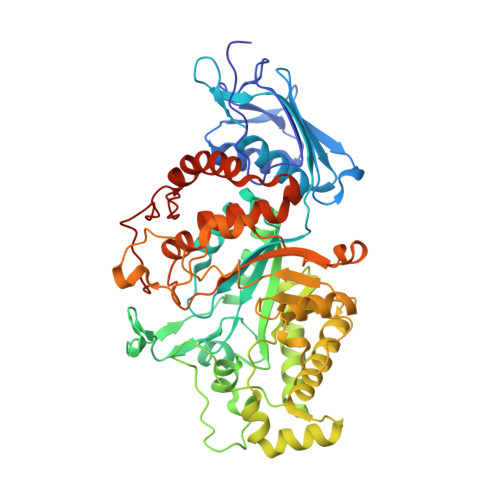 | |
UniProt | |||||
Find proteins for B2UQG6 (Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc)) Explore B2UQG6 Go to UniProtKB: B2UQG6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | B2UQG6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MLA Query on MLA | B [auth A], C [auth A] | MALONIC ACID C3 H4 O4 OFOBLEOULBTSOW-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | D [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 55.584 | α = 90 |
| b = 72.435 | β = 90 |
| c = 137.727 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| MoRDa | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Other government | China | KJ2019ZD02 |