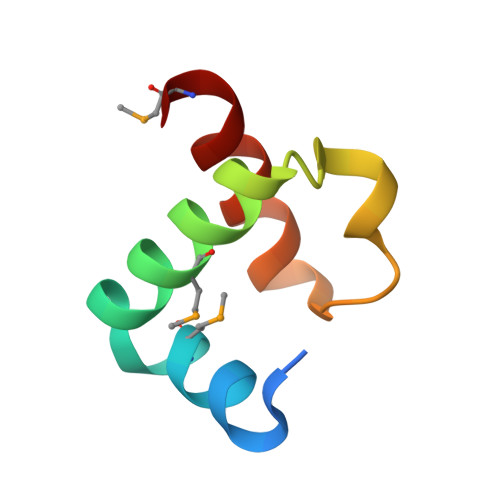
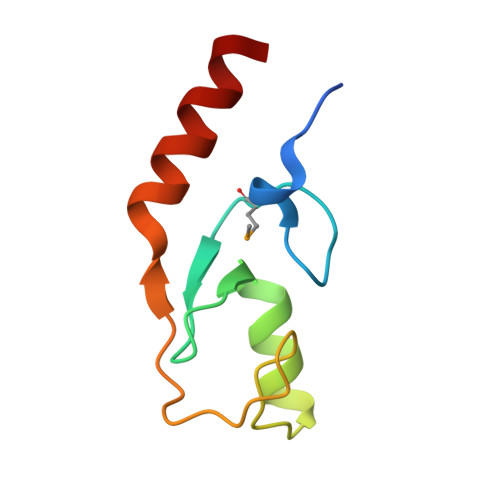
Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection.
Lin, Y., Hu, Q., Zhou, J., Yin, W., Yao, D., Shao, Y., Zhao, Y., Guo, B., Xia, Y., Chen, Q., Wang, Y., Ye, W., Xie, Q., Tyler, B.M., Xing, W., Wang, Y.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33658365
- DOI: https://doi.org/10.1073/pnas.2018312118
- Primary Citation of Related Structures:
7C96 - PubMed Abstract:
Oomycete pathogens such as Phytophthora secrete a repertoire of effectors into host cells to manipulate host immunity and benefit infection. In this study, we found that an RxLR effector, Avr1d, promoted Phytophthora sojae infection in soybean hairy roots. Using a yeast two-hybrid screen, we identified the soybean E3 ubiquitin ligase GmPUB13 as a host target for Avr1d. By coimmunoprecipitation (Co-IP), gel infiltration, and isothermal titration calorimetry (ITC) assays, we confirmed that Avr1d interacts with GmPUB13 both in vivo and in vitro. Furthermore, we found that Avr1d inhibits the E3 ligase activity of GmPUB13. The crystal structure Avr1d in complex with GmPUB13 was solved and revealed that Avr1d occupies the binding site for E2 ubiquitin conjugating enzyme on GmPUB13. In line with this, Avr1d competed with E2 ubiquitin conjugating enzymes for GmPUB13 binding in vitro, thereby decreasing the E3 ligase activity of GmPUB13. Meanwhile, we found that inactivation of the ubiquitin ligase activity of GmPUB13 stabilized GmPUB13 by blocking GmPUB13 degradation. Silencing of GmPUB13 in soybean hairy roots decreased P. sojae infection, suggesting that GmPUB13 acts as a susceptibility factor. Altogether, this study highlights a virulence mechanism of Phytophthora effectors, by which Avr1d competes with E2 for GmPUB13 binding to repress the GmPUB13 E3 ligase activity and thereby stabilizing the susceptibility factor GmPUB13 to facilitate Phytophthora infection. This study unravels the structural basis for modulation of host targets by Phytophthora effectors and will be instrumental for boosting plant resistance breeding.
Organizational Affiliation:
Department of Plant Pathology, Nanjing Agricultural University, 210095 Nanjing, China.