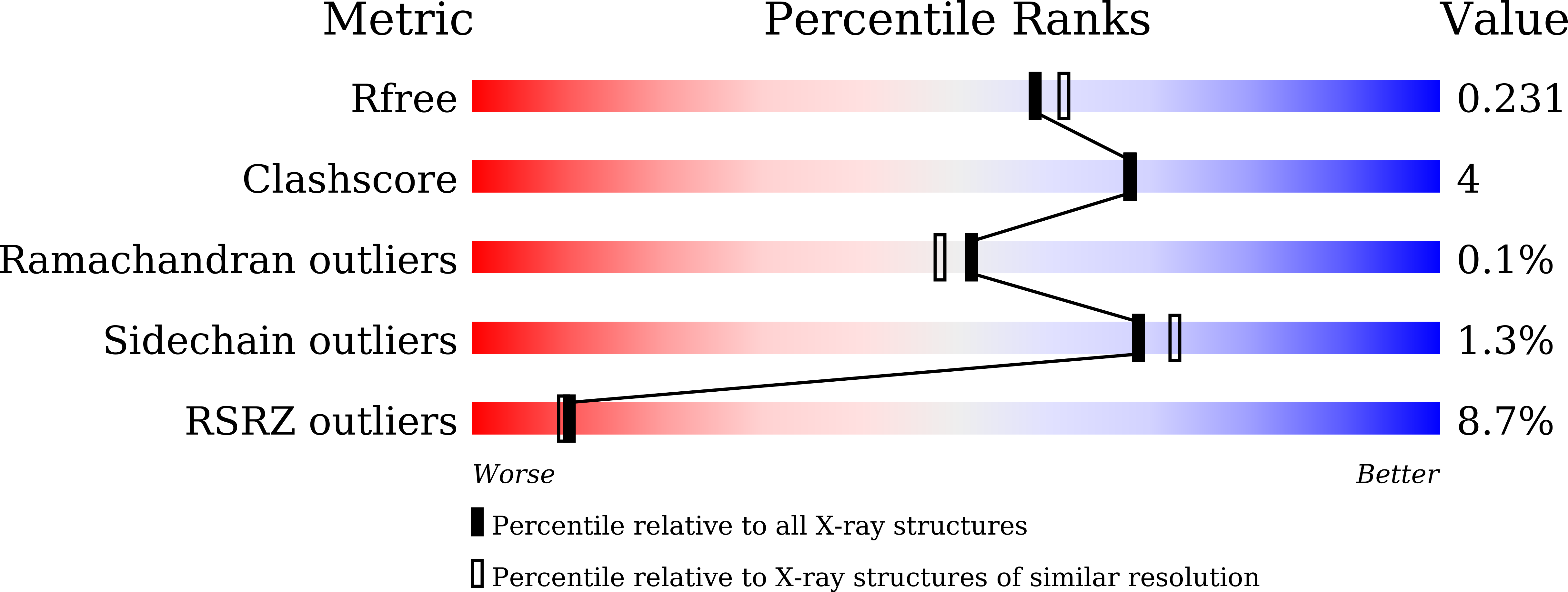
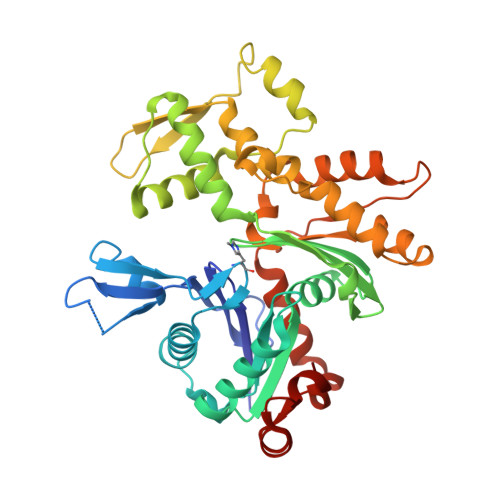
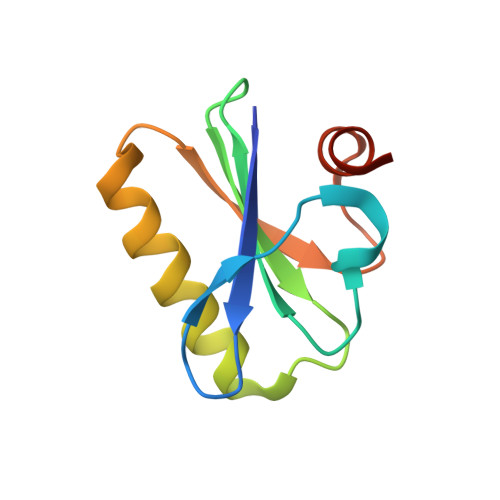
Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea
Akil, C., Tran, L.T., Orhant-Prioux, M., Baskaran, Y., Manser, E., Blanchoin, L., Robinson, R.C.(2020) Proc Natl Acad Sci U S A 117: 19904-19913