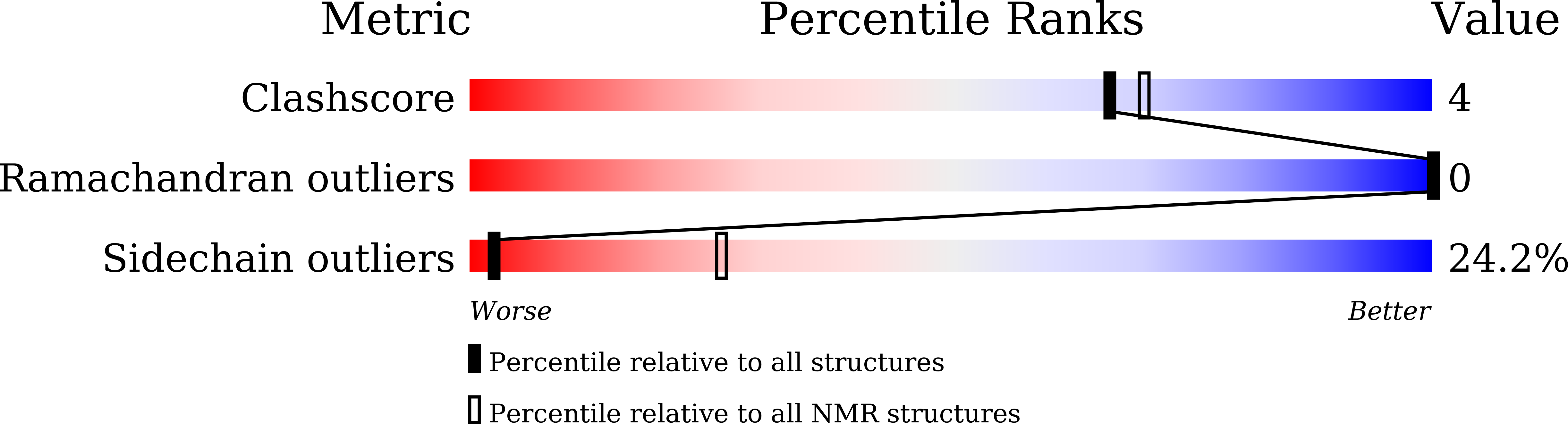Effect of Secondary Structure and Side Chain Length of Hydrophobic Amino Acid Residues on the Antimicrobial Activity and Toxicity of 14-Residue-Long de novo AMPs.
Pandit, G., Chowdhury, N., Abdul Mohid, S., Bidkar, A.P., Bhunia, A., Chatterjee, S.(2021) ChemMedChem 16: 355-367
- PubMed: 33026188
- DOI: https://doi.org/10.1002/cmdc.202000550
- Primary Citation of Related Structures:
7BX2 - PubMed Abstract:
Herein we report the efficacy and toxicity of three de novo designed cationic antimicrobial peptides (AMPs) LL-14, VV-14 and ββ-14, where side chains of the hydrophobic amino acids were reduced gradually. The AMPs showed broad-spectrum antimicrobial activity against three pathogens from the ESKAPE group and two fungal strains. This study showed that side chains which are either too long or too short increase toxicity and lower antimicrobial activity, respectively. VV-14 was found to be non-cytotoxic and highly potent under physiological salt concentrations against several pathogens, especially Salmonella typhi TY2. These AMPs acted via membrane deformation, depolarization, and lysis. The activity of the AMPs is related to their ability to take on amphipathic helical conformations in the presence of microbial membrane mimics. Among AMPs with the same charge, hydrophobic interactions between the side chains of the residues with cell membrane lipids determine their antimicrobial potency and cytotoxicity. Strikingly, an optimum hydrophobic interaction is the crux of generating highly potent non-cytotoxic AMPs.
Organizational Affiliation:
Department of Chemistry, Indian Institute of Technology Guwahati, Guwahati, Assam, India.