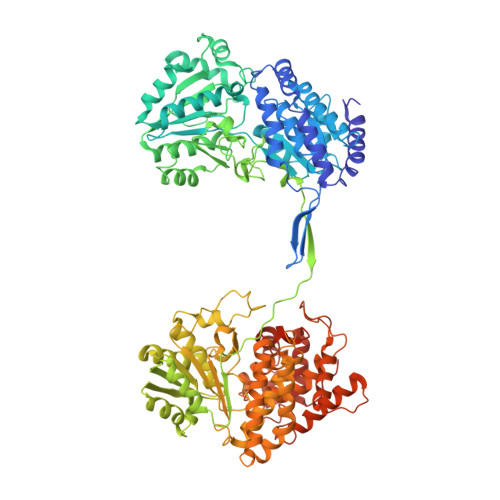
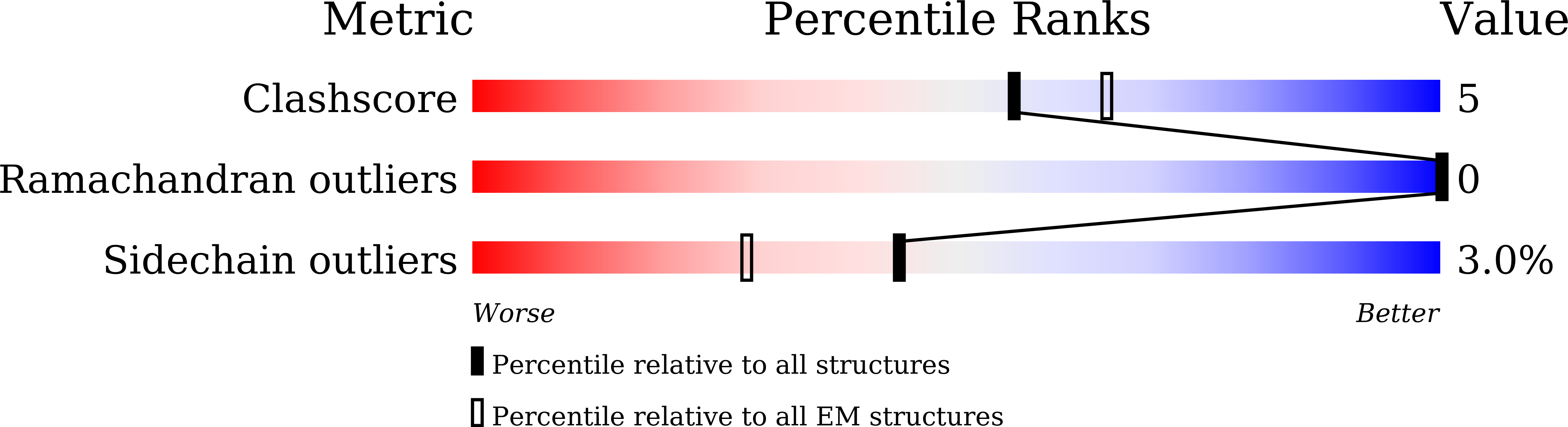Aldehyde-alcohol dehydrogenase undergoes structural transition to form extended spirosomes for substrate channeling.
Kim, G., Yang, J., Jang, J., Choi, J.S., Roe, A.J., Byron, O., Seok, C., Song, J.J.(2020) Commun Biol 3: 298-298
- PubMed: 32523125
- DOI: https://doi.org/10.1038/s42003-020-1030-1
- Primary Citation of Related Structures:
7BVP - PubMed Abstract:
Aldehyde-alcohol dehydrogenase (AdhE) is an enzyme responsible for converting acetyl-CoA to ethanol via acetaldehyde using NADH. AdhE is composed of two catalytic domains of aldehyde dehydrogenase (ALDH) and alcohol dehydrogenase (ADH), and forms a spirosome architecture critical for AdhE activity. Here, we present the atomic resolution (3.43 Å) cryo-EM structure of AdhE spirosomes in an extended conformation. The cryo-EM structure shows that AdhE spirosomes undergo a structural transition from compact to extended forms, which may result from cofactor binding. This transition leads to access to a substrate channel between ALDH and ADH active sites. Furthermore, prevention of this structural transition by crosslinking hampers the activity of AdhE, suggesting that the structural transition is important for AdhE activity. This work provides a mechanistic understanding of the regulation mechanisms of AdhE activity via structural transition, and a platform to modulate AdhE activity for developing antibiotics and for facilitating biofuel production.
Organizational Affiliation:
Department of Biological Sciences, KI for the BioCentury, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, 34141, Korea.