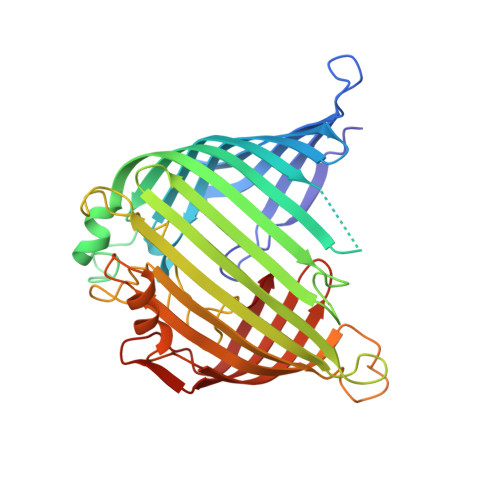
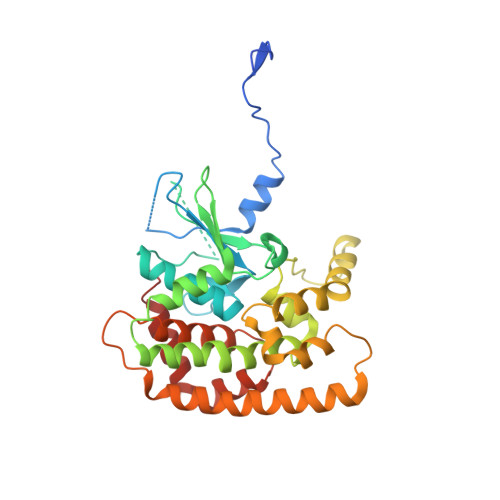
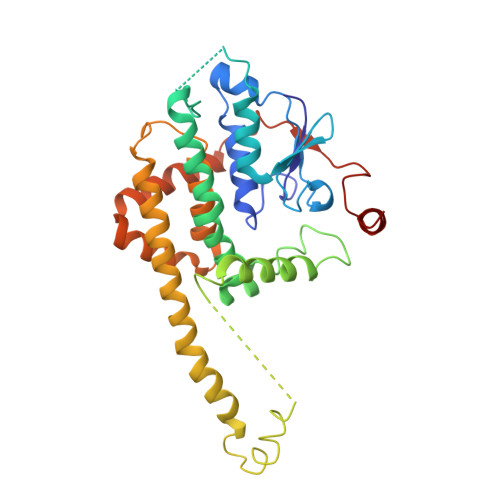
Mitochondrial sorting and assembly machinery operates by beta-barrel switching.
Takeda, H., Tsutsumi, A., Nishizawa, T., Lindau, C., Busto, J.V., Wenz, L.S., Ellenrieder, L., Imai, K., Straub, S.P., Mossmann, W., Qiu, J., Yamamori, Y., Tomii, K., Suzuki, J., Murata, T., Ogasawara, S., Nureki, O., Becker, T., Pfanner, N., Wiedemann, N., Kikkawa, M., Endo, T.(2021) Nature 590: 163-169
- PubMed: 33408415
- DOI: https://doi.org/10.1038/s41586-020-03113-7
- Primary Citation of Related Structures:
7BTW, 7BTX, 7BTY - PubMed Abstract:
The mitochondrial outer membrane contains so-called β-barrel proteins, which allow communication between the cytosol and the mitochondrial interior 1-3 . Insertion of β-barrel proteins into the outer membrane is mediated by the multisubunit mitochondrial sorting and assembly machinery (SAM, also known as TOB) 4-6 . Here we use cryo-electron microscopy to determine the structures of two different forms of the yeast SAM complex at a resolution of 2.8-3.2 Å. The dimeric complex contains two copies of the β-barrel channel protein Sam50-Sam50a and Sam50b-with partially open lateral gates. The peripheral membrane proteins Sam35 and Sam37 cap the Sam50 channels from the cytosolic side, and are crucial for the structural and functional integrity of the dimeric complex. In the second complex, Sam50b is replaced by the β-barrel protein Mdm10. In cooperation with Sam50a, Sam37 recruits and traps Mdm10 by penetrating the interior of its laterally closed β-barrel from the cytosolic side. The substrate-loaded SAM complex contains one each of Sam50, Sam35 and Sam37, but neither Mdm10 nor a second Sam50, suggesting that Mdm10 and Sam50b function as placeholders for a β-barrel substrate released from Sam50a. Our proposed mechanism for dynamic switching of β-barrel subunits and substrate explains how entire precursor proteins can fold in association with the mitochondrial machinery for β-barrel assembly.
Organizational Affiliation:
Faculty of Life Sciences, Kyoto Sangyo University, Kamigamo-motoyama, Kyoto, Japan.