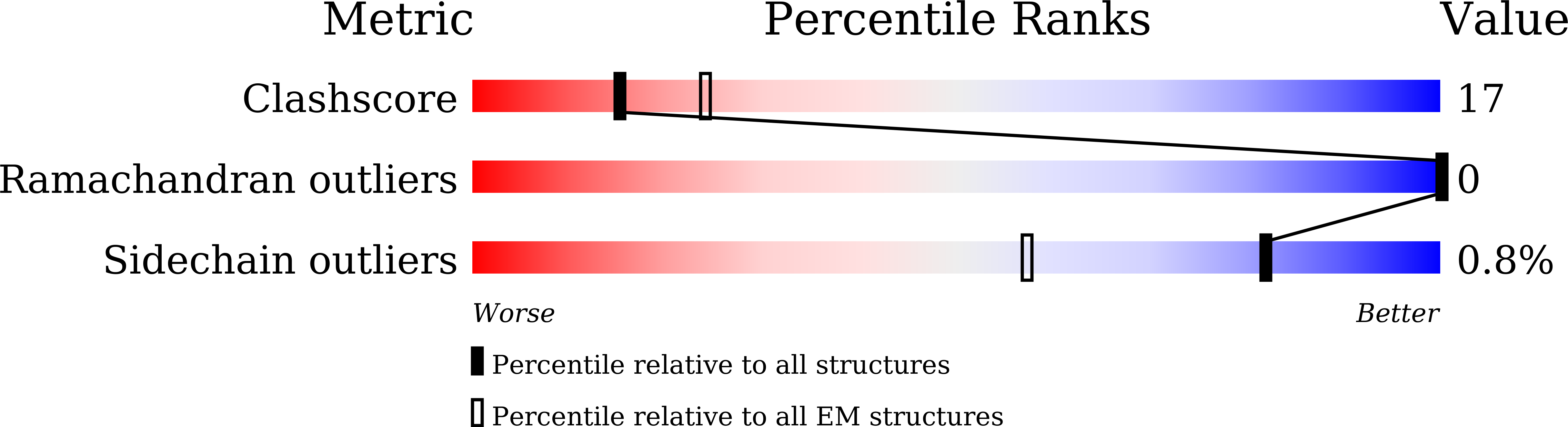Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Zhang, M., Shi, H., Zhang, X., Zhang, X., Huang, Y.(2020) PLoS Biol 18: e3000748-e3000748
- PubMed: 32559189
- DOI: https://doi.org/10.1371/journal.pbio.3000748
- Primary Citation of Related Structures:
6LQH, 6LQJ, 7BRM - PubMed Abstract:
Curli play critical roles in biofilm formation, host cell adhesion, and colonization of inert surfaces in many Enterobacteriaceae. In Escherichia coli, curli biogenesis requires 7 curli-specific gene (csg) products-CsgA through G-working in concert. Of them, CsgG and CsgF are 2 outer membrane (OM)-localized components that consists of the core apparatus for secretion and assembly of curli structural subunits, CsgB and CsgA. Here, we report the cryogenic electron microscopy (cryo-EM) structure of CsgG in complex with CsgF from E. coli. The structure reveals that CsgF forms a stable complex with CsgG via a 1:1 stoichiometry by lining the upper lumen of the nonameric CsgG channel via its N-terminal 27 residues, forming a funnel-like entity plugged in the CsgG channel and creating a unique secretion channel with 2 constriction regions, consistent with the recently reported structure of the CsgG-CsgF complex. Functional studies indicate that export of CsgF to the cell surface requires the CsgG channel, and CsgF not only functions as an adaptor that bridges CsgB with CsgG but also may play important roles in controlling the rates of translocation and/or polymerization for curli structural subunits. Importantly, we found that a series of CsgF-derived peptides are able to efficiently inhibit curli production to E. coli when administrated exogenously, highlighting a potential strategy to interfere biofilm formation in E. coli strains.
Organizational Affiliation:
Shenzhen Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China.