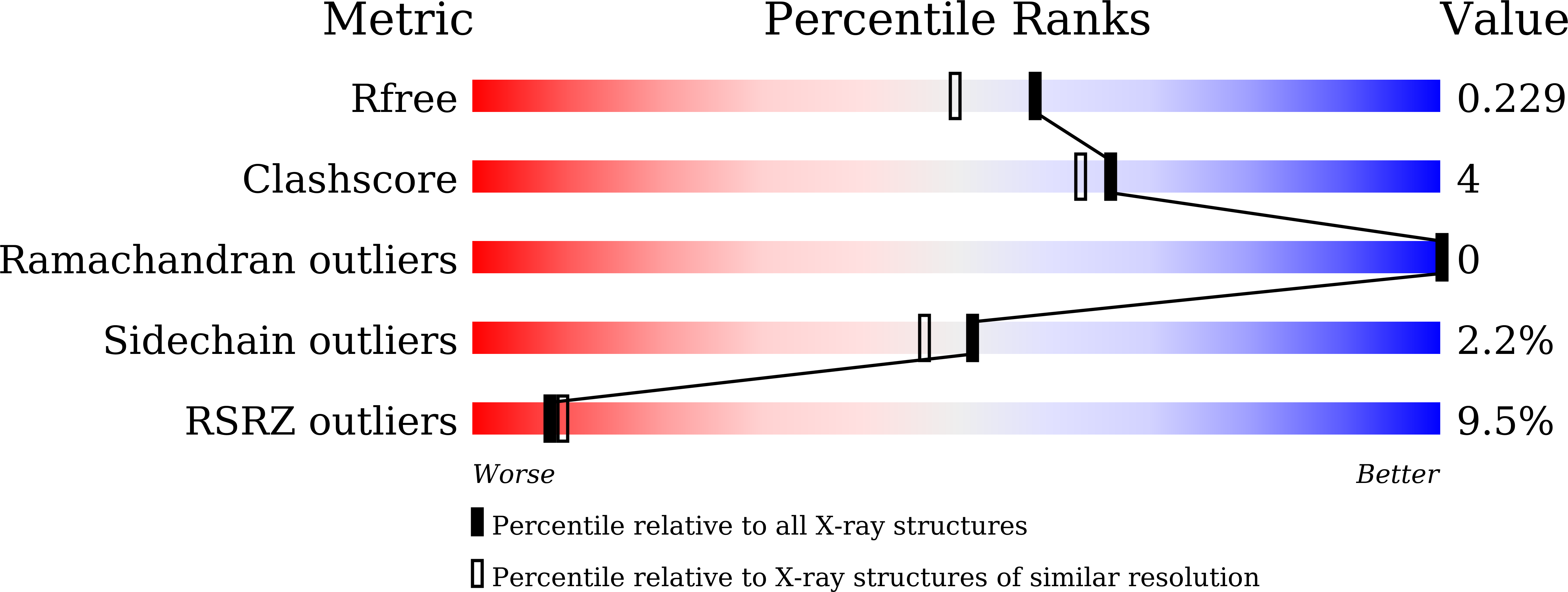
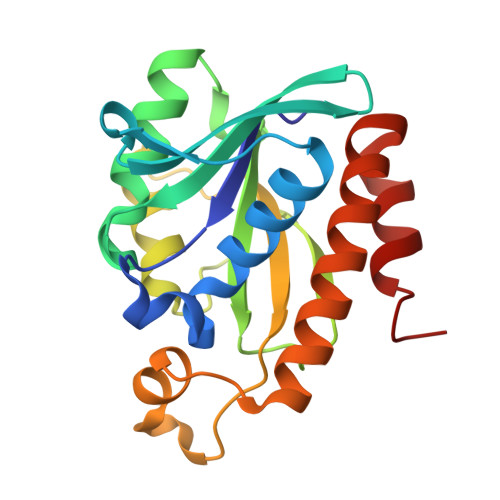
Structural and functional characterization of peptidyl-tRNA hydrolase from Klebsiella pneumoniae.
Mundra, S., Pal, R.K., Tripathi, S., Jain, A., Arora, A.(2021) Biochim Biophys Acta Proteins Proteom 1869: 140554-140554
- PubMed: 33068756
- DOI: https://doi.org/10.1016/j.bbapap.2020.140554
- Primary Citation of Related Structures:
7BRD - PubMed Abstract:
Klebsiella pneumoniae is a member of the ESKAPE panel of pathogens that are top priority to tackle AMR. Bacterial peptidyl tRNA hydrolase (Pth), an essential, ubiquitous enzyme, hydrolyzes the peptidyl-tRNAs that accumulate in the cytoplasm because of premature termination of translation. Pth cleaves the ester bond between 2' or 3' hydroxyl of the ribose in the tRNA and C-terminal carboxylate of the peptide, thereby making free tRNA available for repeated cycles of protein synthesis and preventing cell death by alleviating tRNA starvation. Pth structures have been determined in peptide-bound or peptide-free states. In peptide-bound state, highly conserved residues F67, N69 and N115 adopt a conformation that is conducive to their interaction with peptide moiety of the substrate. While, in peptide-free state, these residues move away from the catalytic center, perhaps, in order to facilitate release of hydrolysed peptide. Here, we present a novel X-ray crystal structure of Pth from Klebsiella pneumoniae (KpPth), at 1.89 Å resolution, in which out of the two molecules in the asymmetric unit, one reflects the peptide-bound while the other reflects peptide-free conformation of the conserved catalytic site residues. Each molecule of the protein has canonical structure with seven stranded β-sheet structure surrounded by six α-helices. MD simulations indicate that both the forms converge over 500 ns simulation to structures with wider opening of the crevice at peptide-binding end. In solution, KpPth is monomeric and its 2D-HSQC spectrum displays a single set of well dispersed peaks. Further, KpPth was demonstrated to be enzymatically active on BODIPY-Lys-tRNA Lys3 .
Organizational Affiliation:
Molecular and Structural Biology Division, CSIR-Central Drug Research Institute, Lucknow 226031, India; Department of Science and Technology, New Delhi 110016, India.