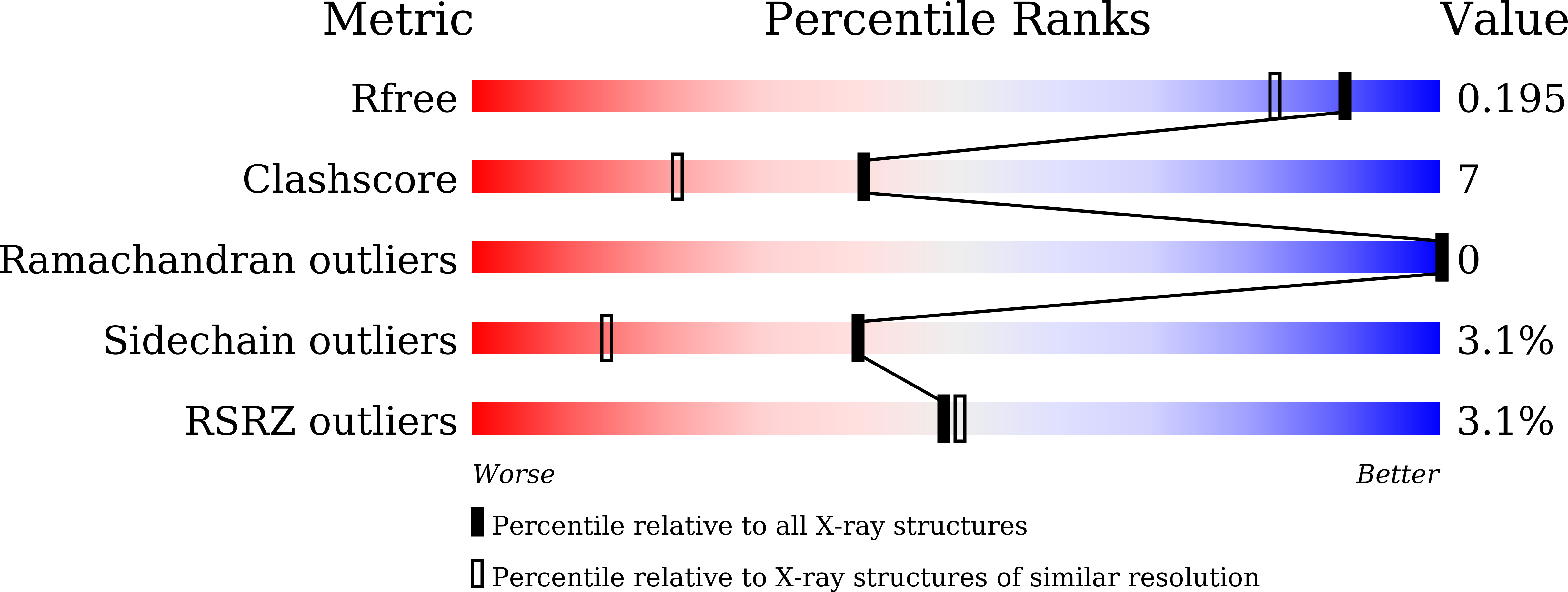
NAP1-Related Protein 1 (NRP1) has multiple interaction modes for chaperoning histones H2A-H2B.
Luo, Q., Wang, B., Wu, Z., Jiang, W., Wang, Y., Du, K., Zhou, N., Zheng, L., Gan, J., Shen, W.H., Ma, J., Dong, A.(2020) Proc Natl Acad Sci U S A 117: 30391-30399
- PubMed: 33199628
- DOI: https://doi.org/10.1073/pnas.2011089117
- Primary Citation of Related Structures:
7BP2, 7BP4, 7BP5, 7BP6, 7C7X - PubMed Abstract:
Nucleosome Assembly Protein 1 (NAP1) family proteins are evolutionarily conserved histone chaperones that play important roles in diverse biological processes. In this study, we determined the crystal structure of Arabidopsis NAP1-Related Protein 1 (NRP1) complexed with H2A-H2B and uncovered a previously unknown interaction mechanism in histone chaperoning. Both in vitro binding and in vivo plant rescue assays proved that interaction mediated by the N-terminal α-helix (αN) domain is essential for NRP1 function. In addition, the C-terminal acidic domain (CTAD) of NRP1 binds to H2A-H2B through a conserved mode similar to other histone chaperones. We further extended previous knowledge of the NAP1-conserved earmuff domain by mapping the amino acids of NRP1 involved in association with H2A-H2B. Finally, we showed that H2A-H2B interactions mediated by αN, earmuff, and CTAD domains are all required for the effective chaperone activity of NRP1. Collectively, our results reveal multiple interaction modes of a NAP1 family histone chaperone and shed light on how histone chaperones shield H2A-H2B from nonspecific interaction with DNA.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, International Associated Laboratory of CNRS-Fudan-HUNAU on Plant Epigenome Research, Department of Biochemistry, Institute of Plant Biology, School of Life Sciences, Fudan University, 200438 Shanghai, China.