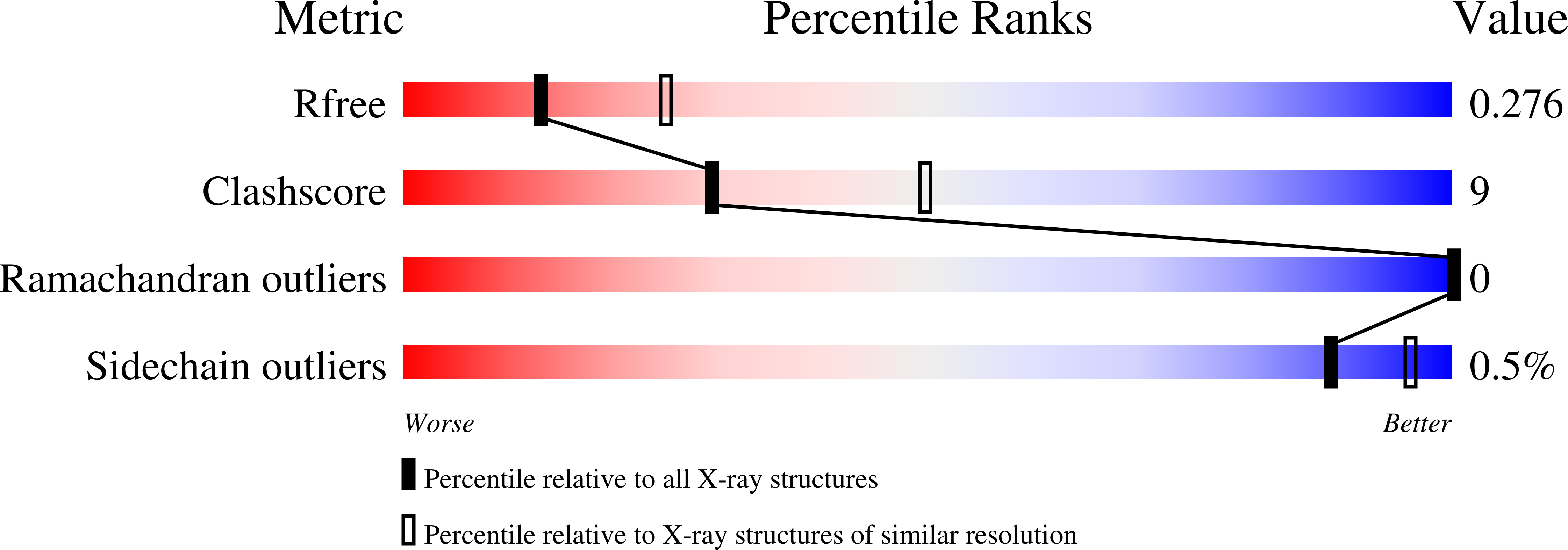
Structural and functional characterization of the bacterial biofilm activator RemA.
Hoffmann, T., Mrusek, D., Bedrunka, P., Burchert, F., Mais, C.N., Kearns, D.B., Altegoer, F., Bremer, E., Bange, G.(2021) Nat Commun 12: 5707-5707
- PubMed: 34588455
- DOI: https://doi.org/10.1038/s41467-021-26005-4
- Primary Citation of Related Structures:
7BM2, 7BME, 7P1W - PubMed Abstract:
Bacillus subtilis can form structurally complex biofilms on solid or liquid surfaces, which requires expression of genes for matrix production. The transcription of these genes is activated by regulatory protein RemA, which binds to poorly conserved, repetitive DNA regions but lacks obvious DNA-binding motifs or domains. Here, we present the structure of the RemA homologue from Geobacillus thermodenitrificans, showing a unique octameric ring with the potential to form a 16-meric superstructure. These results, together with further biochemical and in vivo characterization of B. subtilis RemA, suggests that the protein can wrap DNA around its ring-like structure through a LytTR-related domain.
Organizational Affiliation:
Philipps-University Marburg, Center for Synthetic Microbiology (SYNMIKRO) & Faculty of Biology, Marburg, Germany.