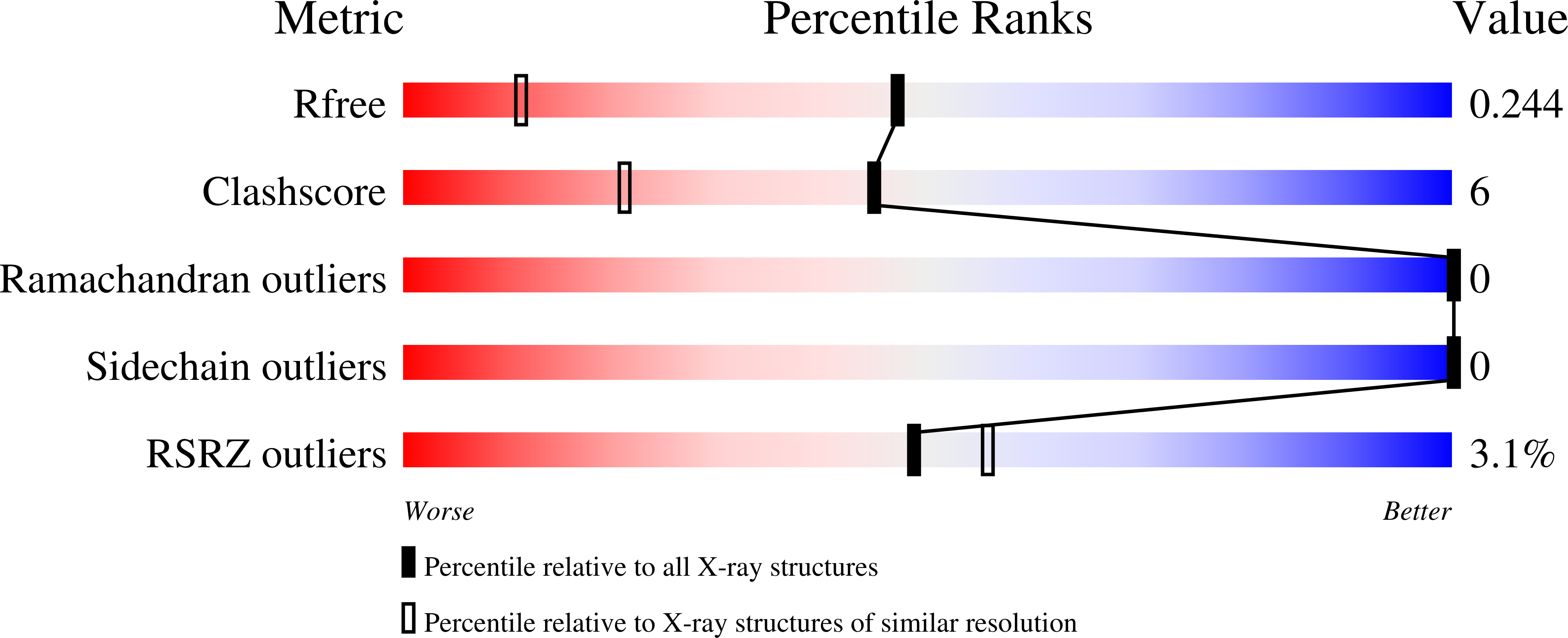
Copper binding and reactivity at the histidine brace motif: insights from mutational analysis of the Pseudomonas fluorescens copper chaperone CopC.
Ipsen, J.O., Hernandez-Rollan, C., Muderspach, S.J., Brander, S., Bertelsen, A.B., Jensen, P.E., Norholm, M.H.H., Lo Leggio, L., Johansen, K.S.(2021) FEBS Lett 595: 1708-1720
- PubMed: 33896006
- DOI: https://doi.org/10.1002/1873-3468.14092
- Primary Citation of Related Structures:
7BK5, 7BK6, 7BK7 - PubMed Abstract:
The histidine brace (His-brace) is a copper-binding motif that is associated with both oxidative enzymes and proteinaceous copper chaperones. Here, we used biochemical and structural methods to characterize mutants of a His-brace-containing copper chaperone from Pseudomonas fluorescens (PfCopC). A total of 15 amino acid variants in primary and second-sphere residues were produced and characterized in terms of their copper binding and redox properties. PfCopC has a very high affinity for Cu(II) and also binds Cu(I). A high reorganization barrier likely prevents redox cycling and, thus, catalysis. In contrast, mutations in the conserved second-sphere Glu27 enable slow oxidation of ascorbate. The crystal structure of the variant E27A confirmed copper binding at the His-brace. Unexpectedly, Asp83 at the equatorial position was shown to be indispensable for Cu(II) binding in the His-brace of PfCopC. A PfCopC mutant that was designed to mimic the His-brace from lytic polysaccharide monooxygenase-like family X325 did not bind Cu(II), but was still able to bind Cu(I). These results highlight the importance of the proteinaceous environment around the copper His-brace for reactivity and, thus, the difference between enzyme and chaperone.
Organizational Affiliation:
Department of Plant and Environmental Sciences, Copenhagen University, Frederiksberg, Denmark.