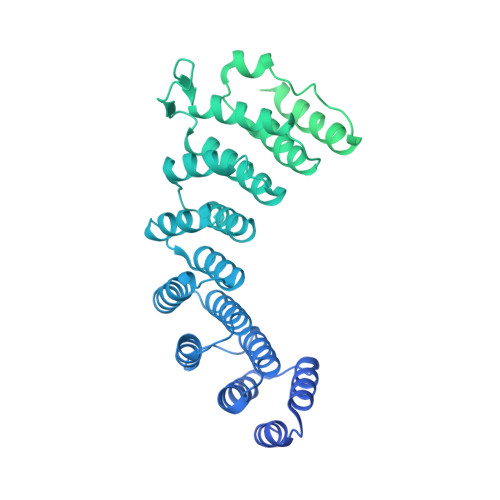
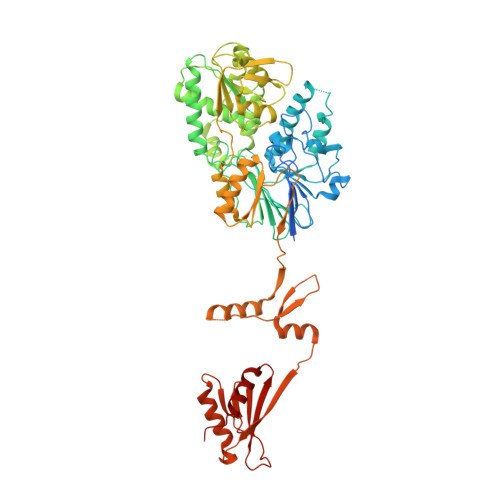
Structure of the catalytic core of the Integrator complex.
Pfleiderer, M.M., Galej, W.P.(2021) Mol Cell 81: 1246
- PubMed: 33548203
- DOI: https://doi.org/10.1016/j.molcel.2021.01.005
- Primary Citation of Related Structures:
7BFP, 7BFQ - PubMed Abstract:
The Integrator is a specialized 3' end-processing complex involved in cleavage and transcription termination of a subset of nascent RNA polymerase II transcripts, including small nuclear RNAs (snRNAs). We provide evidence of the modular nature of the Integrator complex by biochemically characterizing its two subcomplexes, INTS5/8 and INTS10/13/14. Using cryoelectron microscopy (cryo-EM), we determined a 3.5-Å-resolution structure of the INTS4/9/11 ternary complex, which constitutes Integrator's catalytic core. Our structure reveals the spatial organization of the catalytic nuclease INTS11, bound to its catalytically impaired homolog INTS9 via several interdependent interfaces. INTS4, a helical repeat protein, plays a key role in stabilizing nuclease domains and other components. In this assembly, all three proteins form a composite electropositive groove, suggesting a putative RNA binding path within the complex. Comparison with other 3' end-processing machineries points to distinct features and a unique architecture of the Integrator's catalytic module.
Organizational Affiliation:
European Molecular Biology Laboratory, 71 Avenue des Martyrs, 38042 Grenoble, France.