Structural and Biochemical Characterization of a Cold-Active PMGL3 Esterase with Unusual Oligomeric Structure.
Boyko, K.M., Kryukova, M.V., Petrovskaya, L.E., Kryukova, E.A., Nikolaeva, A.Y., Korzhenevsky, D.A., Lomakina, G.Y., Novototskaya-Vlasova, K.A., Rivkina, E.M., Dolgikh, D.A., Kirpichnikov, M.P., Popov, V.O.(2021) Biomolecules 11
- PubMed: 33466452
- DOI: https://doi.org/10.3390/biom11010057
- Primary Citation of Related Structures:
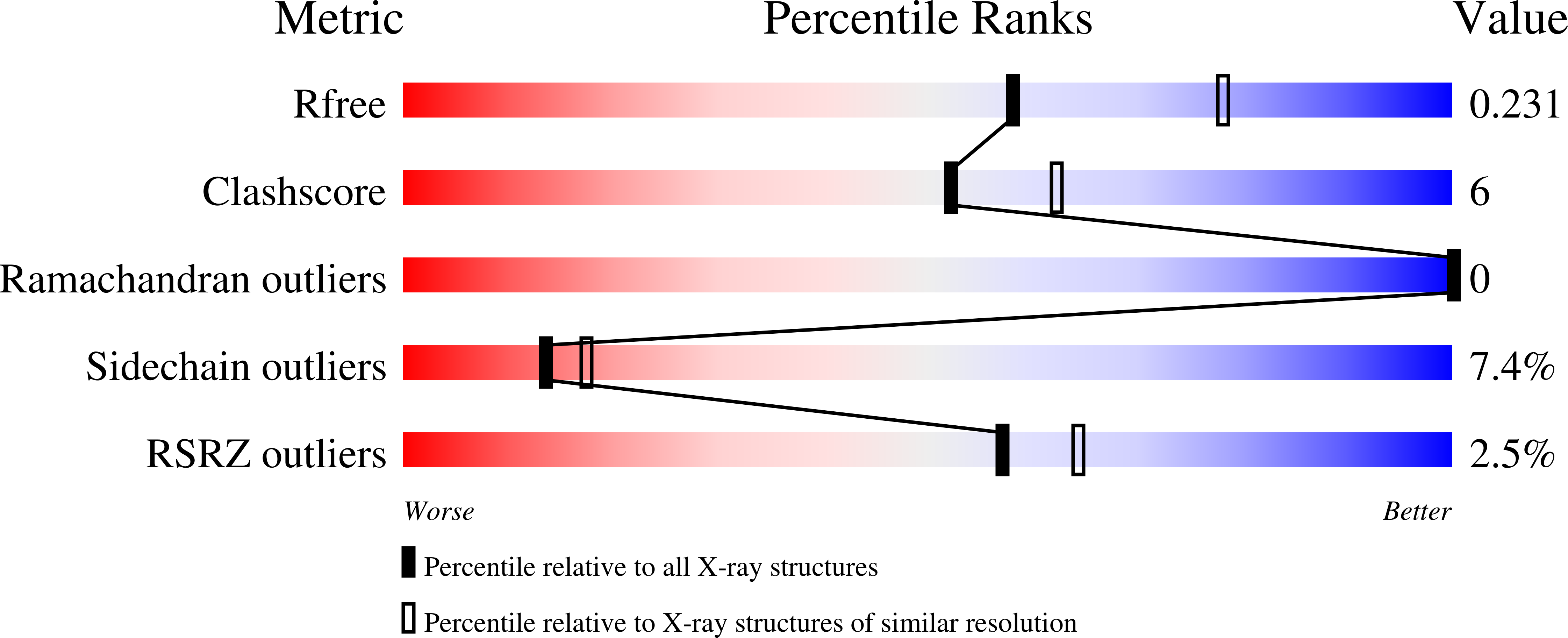
7B1X - PubMed Abstract:
The gene coding for a novel cold-active esterase PMGL3 was previously obtained from a Siberian permafrost metagenomic DNA library and expressed in Escherichia coli . We elucidated the 3D structure of the enzyme which belongs to the hormone-sensitive lipase (HSL) family. Similar to other bacterial HSLs, PMGL3 shares a canonical α/β hydrolase fold and is presumably a dimer in solution but, in addition to the dimer, it forms a tetrameric structure in a crystal and upon prolonged incubation at 4 °C. Detailed analysis demonstrated that the crystal tetramer of PMGL3 has a unique architecture compared to other known tetramers of the bacterial HSLs. To study the role of the specific residues comprising the tetramerization interface of PMGL3, several mutant variants were constructed. Size exclusion chromatography (SEC) analysis of D7N, E47Q, and K67A mutants demonstrated that they still contained a portion of tetrameric form after heat treatment, although its amount was significantly lower in D7N and K67A compared to the wild type. Moreover, the D7N and K67A mutants demonstrated a 40 and 60% increase in the half-life at 40 °C in comparison with the wild type protein. K m values of these mutants were similar to that of the wt PMGL3. However, the catalytic constants of the E47Q and K67A mutants were reduced by ~40%.
Organizational Affiliation:
Bach Institute of Biochemistry, Research Center of Biotechnology of the Russian Academy of Sciences, 119071 Moscow, Russia.