M3258 Is a Selective Inhibitor of the Immunoproteasome Subunit LMP7 ( beta 5i) Delivering Efficacy in Multiple Myeloma Models.
Sanderson, M.P., Friese-Hamim, M., Walter-Bausch, G., Busch, M., Gaus, S., Musil, D., Rohdich, F., Zanelli, U., Downey-Kopyscinski, S.L., Mitsiades, C.S., Schadt, O., Klein, M., Esdar, C.(2021) Mol Cancer Ther 20: 1378-1387
- PubMed: 34045234
- DOI: https://doi.org/10.1158/1535-7163.MCT-21-0005
- Primary Citation of Related Structures:
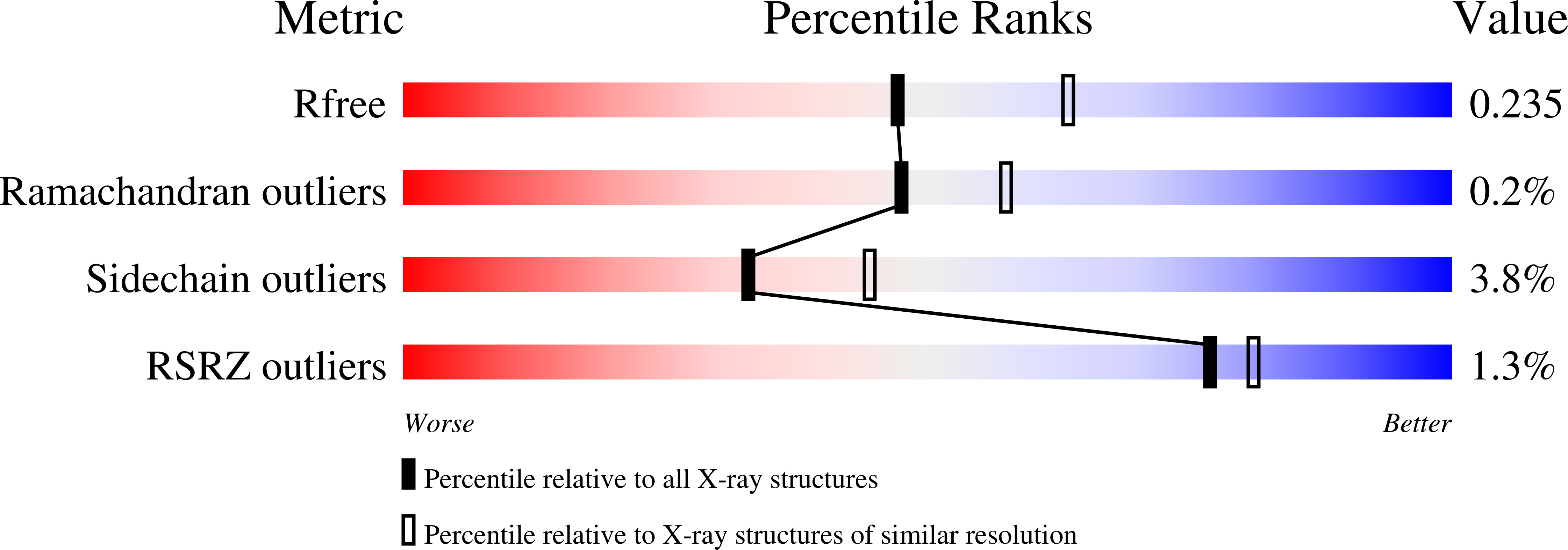
7AWE - PubMed Abstract:
Large multifunctional peptidase 7 (LMP7/β5i/PSMB8) is a proteolytic subunit of the immunoproteasome, which is predominantly expressed in normal and malignant hematolymphoid cells, including multiple myeloma, and contributes to the degradation of ubiquitinated proteins. Described herein for the first time is the preclinical profile of M3258; an orally bioavailable, potent, reversible and highly selective LMP7 inhibitor. M3258 demonstrated strong antitumor efficacy in multiple myeloma xenograft models, including a novel model of the human bone niche of multiple myeloma. M3258 treatment led to a significant and prolonged suppression of tumor LMP7 activity and ubiquitinated protein turnover and the induction of apoptosis in multiple myeloma cells both in vitro and in vivo Furthermore, M3258 showed superior antitumor efficacy in selected multiple myeloma and mantle cell lymphoma xenograft models compared with the approved nonselective proteasome inhibitors bortezomib and ixazomib. The differentiated preclinical profile of M3258 supported the initiation of a phase I study in patients with multiple myeloma (NCT04075721).
Organizational Affiliation:
Merck KGaA, Darmstadt, Germany. michael.sanderson@merckgroup.com.