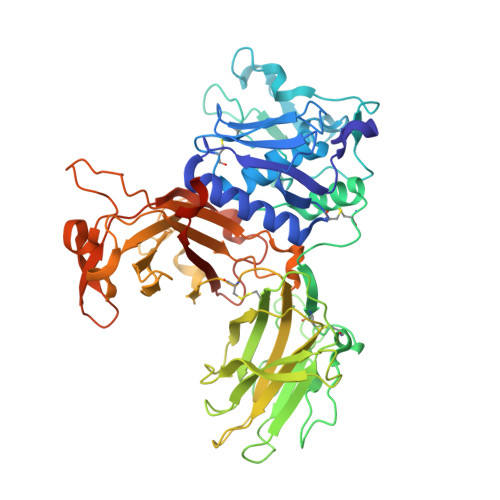
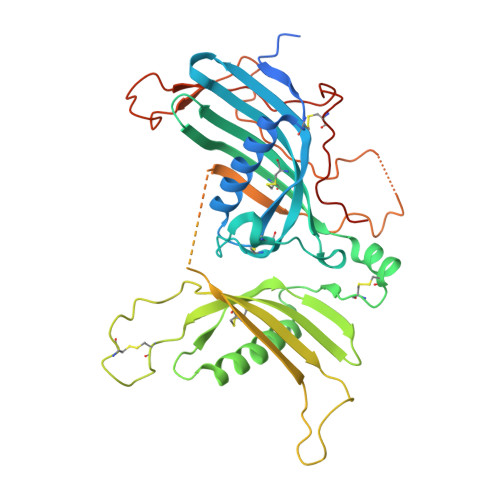
The crystal structure of a 250-kDa heterotetrameric particle explains inhibition of sheddase meprin beta by endogenous fetuin-B.
Eckhard, U., Korschgen, H., von Wiegen, N., Stocker, W., Gomis-Ruth, F.X.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33782129
- DOI: https://doi.org/10.1073/pnas.2023839118
- Primary Citation of Related Structures:
7AUW - PubMed Abstract:
Meprin β (Mβ) is a multidomain type-I membrane metallopeptidase that sheds membrane-anchored substrates, releasing their soluble forms. Fetuin-B (FB) is its only known endogenous protein inhibitor. Herein, we analyzed the interaction between the ectodomain of Mβ (MβΔC) and FB, which stabilizes the enzyme and inhibits it with subnanomolar affinity. The MβΔC:FB crystal structure reveals a ∼250-kDa, ∼160-Å polyglycosylated heterotetrameric particle with a remarkable glycan structure. Two FB moieties insert like wedges through a "CPDCP trunk" and two hairpins into the respective peptidase catalytic domains, blocking the catalytic zinc ions through an "aspartate switch" mechanism. Uniquely, the active site clefts are obstructed from subsites S 4 to S 10 ', but S 1 and S 1 ' are spared, which prevents cleavage. Modeling of full-length Mβ reveals an EGF-like domain between MβΔC and the transmembrane segment that likely serves as a hinge to transit between membrane-distal and membrane-proximal conformations for inhibition and catalysis, respectively.
Organizational Affiliation:
Proteolysis Laboratory, Department of Structural Biology, Molecular Biology Institute of Barcelona Higher Scientific Research Council, Barcelona Science Park, 08028 Barcelona, Spain.