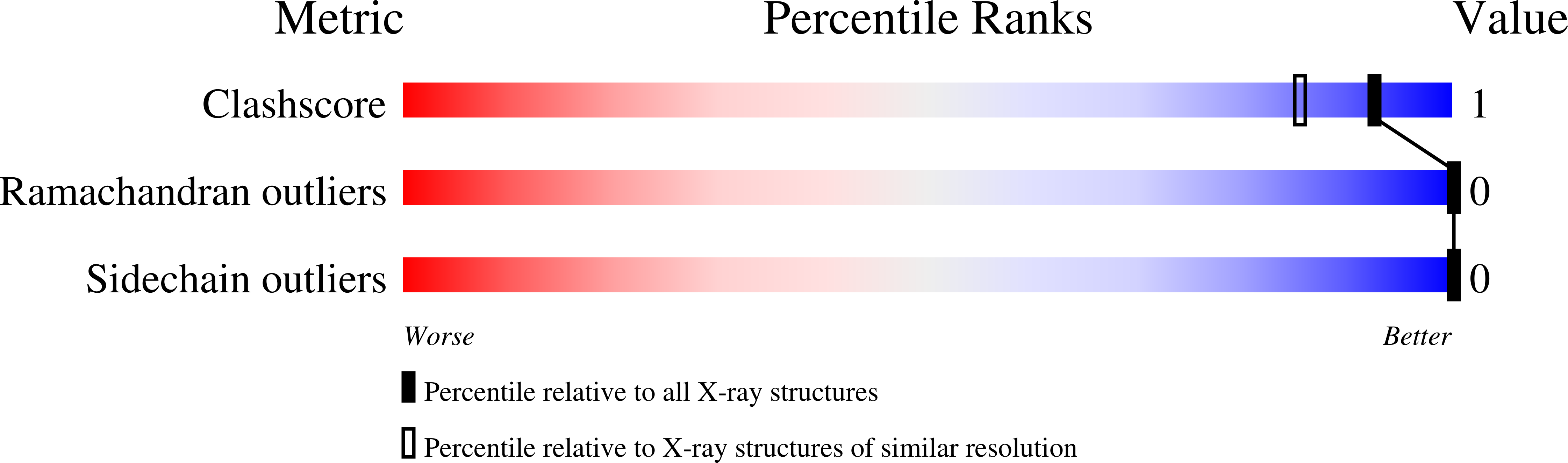Molecular Plasticity of Crystalline CK2 alpha ' Leads to KN2, a Bivalent Inhibitor of Protein Kinase CK2 with Extraordinary Selectivity.
Lindenblatt, D., Applegate, V., Nickelsen, A., Klussmann, M., Neundorf, I., Gotz, C., Jose, J., Niefind, K.(2022) J Med Chem 65: 1302-1312
- PubMed: 34323071
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00063
- Primary Citation of Related Structures:
7AT5, 7AT9, 7ATV - PubMed Abstract:
CK2α and CK2α' are paralogous catalytic subunits of CK2, which belongs to the eukaryotic protein kinases. CK2 promotes tumorigenesis and the spread of pathogenic viruses like SARS-CoV-2 and is thus an attractive drug target. Efforts to develop selective CK2 inhibitors binding offside the ATP site had disclosed the αD pocket in CK2α; its occupation requires large conformational adaptations of the helix αD. As shown here, the αD pocket is accessible also in CK2α', where the necessary structural plasticity can be triggered with suitable ligands even in the crystalline state. A CK2α' structure with an ATP site and an αD pocket ligand guided the design of the bivalent CK2 inhibitor KN2. It binds to CK2 with low nanomolar affinity, is cell-permeable, and suppresses the intracellular phosphorylation of typical CK2 substrates. Kinase profiling revealed a high selectivity of KN2 for CK2 and emphasizes the selectivity-promoting potential of the αD pocket.
Organizational Affiliation:
Department für Chemie, Institut für Biochemie, Universität zu Köln, Zülpicher Str. 47, D-50674 Köln, Germany.