Plant-expressed virus-like particles reveal the intricate maturation process of a eukaryotic virus.
Castells-Graells, R., Ribeiro, J.R.S., Domitrovic, T., Hesketh, E.L., Scarff, C.A., Johnson, J.E., Ranson, N.A., Lawson, D.M., Lomonossoff, G.P.(2021) Commun Biol 4: 619-619
- PubMed: 34031522
- DOI: https://doi.org/10.1038/s42003-021-02134-w
- Primary Citation of Related Structures:
7ANM, 7ATA - PubMed Abstract:
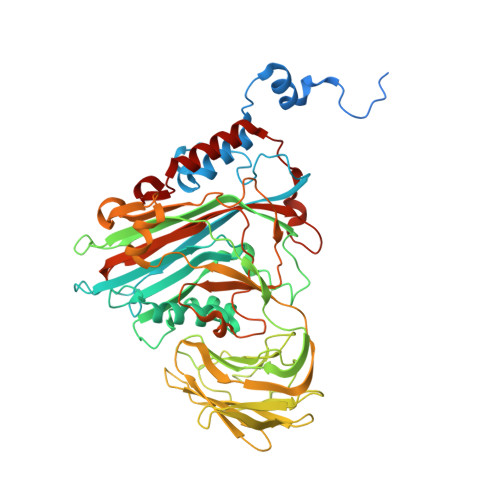
Many virus capsids undergo exquisitely choreographed maturation processes in their host cells to produce infectious virions, and these remain poorly understood. As a tool for studying virus maturation, we transiently expressed the capsid protein of the insect virus Nudaurelia capensis omega virus (NωV) in Nicotiana benthamiana and were able to purify both immature procapsids and mature capsids from infiltrated leaves by varying the expression time. Cryo-EM analysis of the plant-produced procapsids and mature capsids to 6.6 Å and 2.7 Å resolution, respectively, reveals that in addition to large scale rigid body motions, internal regions of the subunits are extensively remodelled during maturation, creating the active site required for autocatalytic cleavage and infectivity. The mature particles are biologically active in terms of their ability to lyse membranes and have a structure that is essentially identical to authentic virus. The ability to faithfully recapitulate and visualize a complex maturation process in plants, including the autocatalytic cleavage of the capsid protein, has revealed a ~30 Å translation-rotation of the subunits during maturation as well as conformational rearrangements in the N and C-terminal helical regions of each subunit.
Organizational Affiliation:
Department of Biological Chemistry, John Innes Centre, Colney, UK.