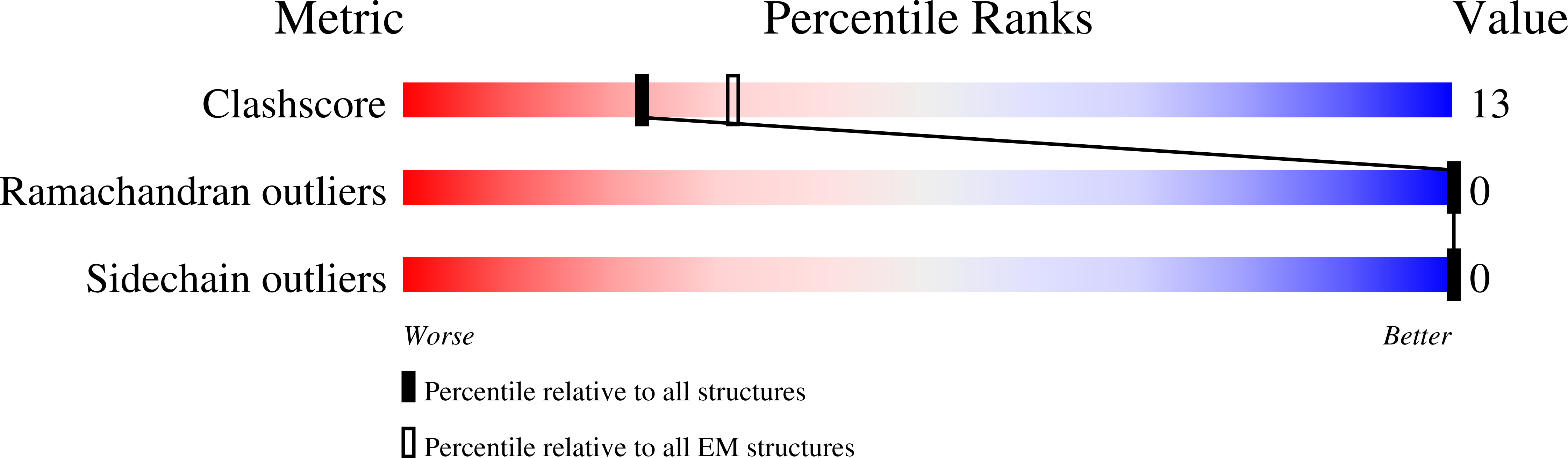Capsid opening enables genome release of iflaviruses.
Skubnik, K., Sukenik, L., Buchta, D., Fuzik, T., Prochazkova, M., Moravcova, J., Smerdova, L., Pridal, A., Vacha, R., Plevka, P.(2021) Sci Adv 7
- PubMed: 33523856
- DOI: https://doi.org/10.1126/sciadv.abd7130
- Primary Citation of Related Structures:
7AL3 - PubMed Abstract:
The family Iflaviridae includes economically important viruses of the western honeybee such as deformed wing virus, slow bee paralysis virus, and sacbrood virus. Iflaviruses have nonenveloped virions and capsids organized with icosahedral symmetry. The genome release of iflaviruses can be induced in vitro by exposure to acidic pH, implying that they enter cells by endocytosis. Genome release intermediates of iflaviruses have not been structurally characterized. Here, we show that conformational changes and expansion of iflavirus RNA genomes, which are induced by acidic pH, trigger the opening of iflavirus particles. Capsids of slow bee paralysis virus and sacbrood virus crack into pieces. In contrast, capsids of deformed wing virus are more flexible and open like flowers to release their genomes. The large openings in iflavirus particles enable the fast exit of genomes from capsids, which decreases the probability of genome degradation by the RNases present in endosomes.
Organizational Affiliation:
Central European Institute of Technology, Masaryk University, Kamenice 753/5, 625 00 Brno, Czech Republic.