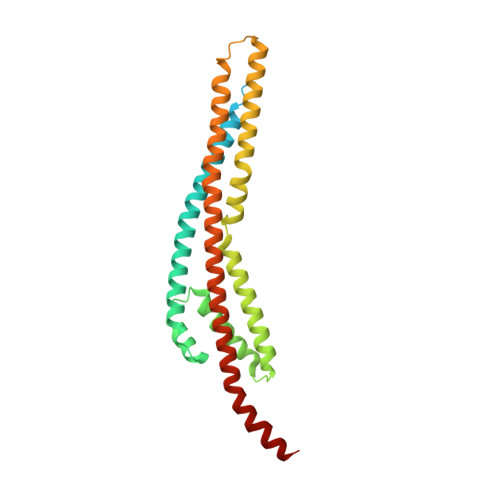
Structural and molecular determinants for the interaction of ExbB from Serratia marcescens and HasB, a TonB paralog.
Biou, V., Adaixo, R.J.D., Chami, M., Coureux, P.D., Laurent, B., Enguene, V.Y.N., de Amorim, G.C., Izadi-Pruneyre, N., Malosse, C., Chamot-Rooke, J., Stahlberg, H., Delepelaire, P.(2022) Commun Biol 5: 355-355
- PubMed: 35418619
- DOI: https://doi.org/10.1038/s42003-022-03306-y
- Primary Citation of Related Structures:
6YE4, 7AJQ - PubMed Abstract:
ExbB and ExbD are cytoplasmic membrane proteins that associate with TonB to convey the energy of the proton-motive force to outer membrane receptors in Gram-negative bacteria for iron uptake. The opportunistic pathogen Serratia marcescens (Sm) possesses both TonB and a heme-specific TonB paralog, HasB. ExbB Sm has a long periplasmic extension absent in other bacteria such as E. coli (Ec). Long ExbB's are found in several genera of Alphaproteobacteria, most often in correlation with a hasB gene. We investigated specificity determinants of ExbB Sm and HasB. We determined the cryo-EM structures of ExbB Sm and of the ExbB-ExbD Sm complex from S. marcescens. ExbB Sm alone is a stable pentamer, and its complex includes two ExbD monomers. We showed that ExbB Sm extension interacts with HasB and is involved in heme acquisition and we identified key residues in the membrane domain of ExbB Sm and ExbB Ec , essential for function and likely involved in the interaction with TonB/HasB. Our results shed light on the class of inner membrane energy machinery formed by ExbB, ExbD and HasB.
Organizational Affiliation:
Laboratoire de Biologie Physico-Chimique des Protéines Membranaires, Université de Paris, UMR 7099 CNRS, F-75005, Paris, France. valerie.biou@ibpc.fr.