Structure and metal-binding properties of PA4063, a novel player in periplasmic zinc trafficking by Pseudomonas aeruginosa.
Fiorillo, A., Battistoni, A., Ammendola, S., Secli, V., Rinaldo, S., Cutruzzola, F., Demitri, N., Ilari, A.(2021) Acta Crystallogr D Struct Biol 77: 1401-1410
- PubMed: 34726168
- DOI: https://doi.org/10.1107/S2059798321009608
- Primary Citation of Related Structures:
7AHW, 7ALY, 7AMX, 7BGO - PubMed Abstract:
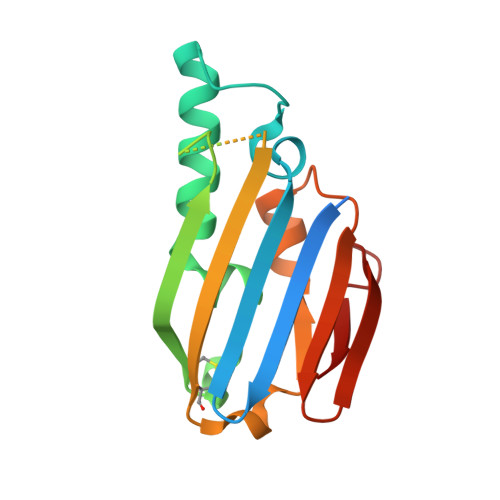
The capability to obtain essential nutrients in hostile environments is a critical skill for pathogens. Under zinc-deficient conditions, Pseudomonas aeruginosa expresses a pool of metal homeostasis control systems that is complex compared with other Gram-negative bacteria and has only been partially characterized. Here, the structure and zinc-binding properties of the protein PA4063, the first component of the PA4063-PA4066 operon, are described. PA4063 has no homologs in other organisms and is characterized by the presence of two histidine-rich sequences. ITC titration detected two zinc-binding sites with micromolar affinity. Crystallographic characterization, performed both with and without zinc, revealed an α/β-sandwich structure that can be classified as a noncanonical ferredoxin-like fold since it differs in size and topology. The histidine-rich stretches located at the N-terminus and between β3 and β4 are disordered in the apo structure, but a few residues become structured in the presence of zinc, contributing to coordination in one of the two sites. The ability to bind two zinc ions at relatively low affinity, the absence of catalytic cavities and the presence of two histidine-rich loops are properties and structural features which suggest that PA4063 might play a role as a periplasmic zinc chaperone or as a concentration sensor useful for optimizing the response of the pathogen to zinc deficiency.
Organizational Affiliation:
Department of Biochemical Sciences, Sapienza University of Rome, Pizzale Aldo Moro 5, 00185 Rome, Italy.