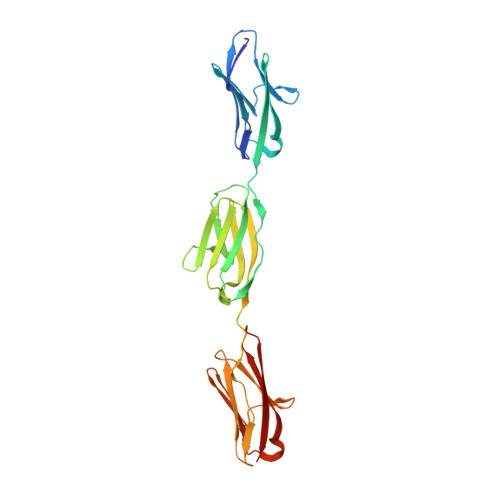
The N2A region of titin has a unique structural configuration.
Stronczek, C., Lange, S., Bullard, B., Wolniak, S., Borgeson, E., Mayans, O., Fleming, J.R.(2021) J Gen Physiol 153
- PubMed: 33836065
- DOI: https://doi.org/10.1085/jgp.202012766
- Primary Citation of Related Structures:
7AHS - PubMed Abstract:
The N2A segment of titin is a main signaling hub in the sarcomeric I-band that recruits various signaling factors and processing enzymes. It has also been proposed to play a role in force production through its Ca2+-regulated association with actin. However, the molecular basis by which N2A performs these functions selectively within the repetitive and extensive titin chain remains poorly understood. Here, we analyze the structure of N2A components and their association with F-actin. Specifically, we characterized the structure of its Ig domains by elucidating the atomic structure of the I81-I83 tandem using x-ray crystallography and computing a homology model for I80. Structural data revealed these domains to present heterogeneous and divergent Ig folds, where I81 and I83 have unique loop structures. Notably, the I81-I83 tandem has a distinct rotational chain arrangement that confers it a unique multi-domain topography. However, we could not identify specific Ca2+-binding sites in these Ig domains, nor evidence of the association of titin N2A components with F-actin in transfected C2C12 myoblasts or C2C12-derived myotubes. In addition, F-actin cosedimentation assays failed to reveal binding to N2A. We conclude that N2A has a unique architecture that predictably supports its selective recruitment of binding partners in signaling, but that its mechanical role through interaction with F-actin awaits validation.
Organizational Affiliation:
Department of Biology, University of Konstanz, Konstanz, Germany.