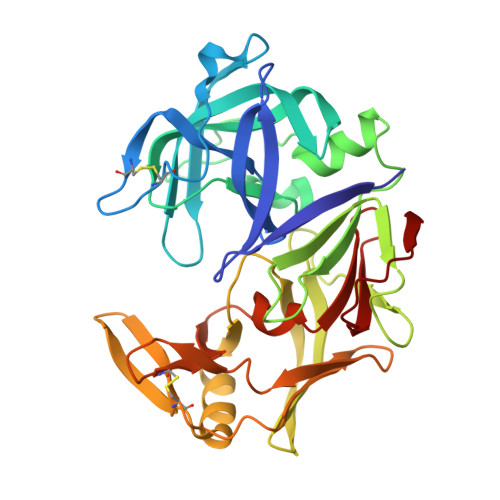
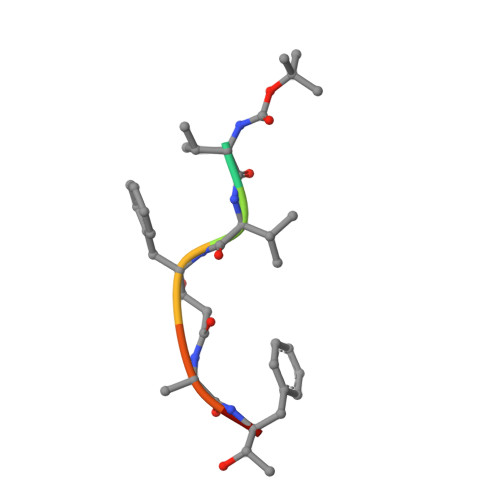
Structural determinants for subnanomolar inhibition of the secreted aspartic protease Sapp1p from Candida parapsilosis .
Dostal, J., Brynda, J., Vankova, L., Zia, S.R., Pichova, I., Heidingsfeld, O., Lepsik, M.(2021) J Enzyme Inhib Med Chem 36: 914-921
- PubMed: 33843395
- DOI: https://doi.org/10.1080/14756366.2021.1906664
- Primary Citation of Related Structures:
7AGB, 7AGC, 7AGD, 7AGE - PubMed Abstract:
Pathogenic Candida albicans yeasts frequently cause infections in hospitals. Antifungal drugs lose effectiveness due to other Candida species and resistance. New medications are thus required. Secreted aspartic protease of C. parapsilosis (Sapp1p) is a promising target. We have thus solved the crystal structures of Sapp1p complexed to four peptidomimetic inhibitors. Three potent inhibitors (K i : 0.1, 0.4, 6.6 nM) resembled pepstatin A (K i : 0.3 nM), a general aspartic protease inhibitor, in terms of their interactions with Sapp1p. However, the weaker inhibitor (K i : 14.6 nM) formed fewer nonpolar contacts with Sapp1p, similarly to the smaller HIV protease inhibitor ritonavir (K i : 1.9 µM), which, moreover, formed fewer H-bonds. The analyses have revealed the structural determinants of the subnanomolar inhibition of C. parapsilosis aspartic protease. Because of the high similarity between Saps from different Candida species, these results can further be used for the design of potent and specific Sap inhibitor-based antimycotic drugs.
Organizational Affiliation:
Institute of Organic Chemistry and Biochemistry, Czech Academy of Sciences, Prague, Czech Republic.