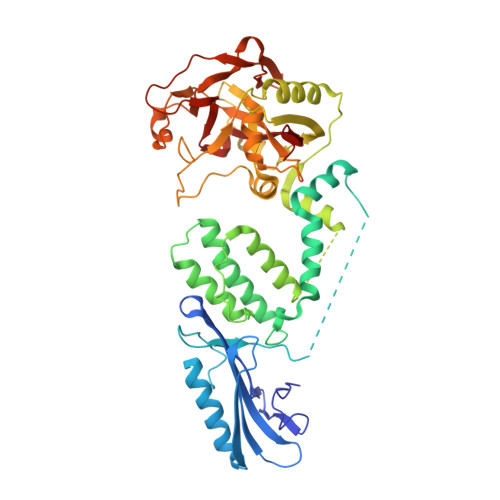
Activation of PARP2/ARTD2 by DNA damage induces conformational changes relieving enzyme autoinhibition.
Obaji, E., Maksimainen, M.M., Galera-Prat, A., Lehtio, L.(2021) Nat Commun 12: 3479-3479
- PubMed: 34108479
- DOI: https://doi.org/10.1038/s41467-021-23800-x
- Primary Citation of Related Structures:
7AEO - PubMed Abstract:
Human PARP2/ARTD2 is an ADP-ribosyltransferase which, when activated by 5'-phosphorylated DNA ends, catalyses poly-ADP-ribosylation of itself, other proteins and DNA. In this study, a crystal structure of PARP2 in complex with an activating 5'-phosphorylated DNA shows that the WGR domain bridges the dsDNA gap and joins the DNA ends. This DNA binding results in major conformational changes, including reorganization of helical fragments, in the PARP2 regulatory domain. A comparison of PARP1 and PARP2 crystal structures reveals how binding to a DNA damage site leads to formation of a catalytically competent conformation. In this conformation, PARP2 is capable of binding substrate NAD + and histone PARylation factor 1 that changes PARP2 residue specificity from glutamate to serine when initiating DNA repair processes. The structure also reveals how the conformational changes in the autoinhibitory regulatory domain would promote the flexibility needed by the enzyme to reach the target macromolecule for ADP-ribosylation.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Medicine & Biocenter Oulu, University of Oulu, Oulu, Finland.