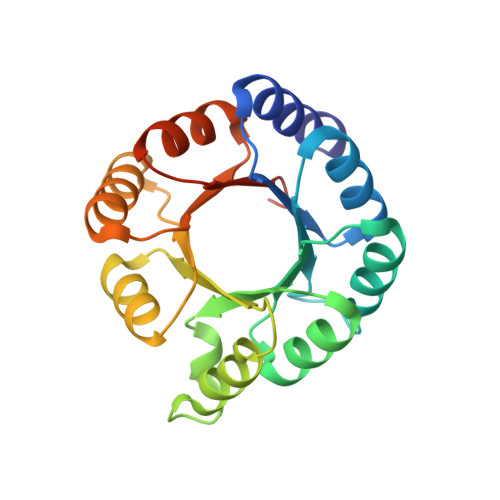
Extension of a de novo TIM barrel with a rationally designed secondary structure element.
Wiese, J.G., Shanmugaratnam, S., Hocker, B.(2021) Protein Sci 30: 982-989
- PubMed: 33723882
- DOI: https://doi.org/10.1002/pro.4064
- Primary Citation of Related Structures:
7A8S - PubMed Abstract:
The ability to construct novel enzymes is a major aim in de novo protein design. A popular enzyme fold for design attempts is the TIM barrel. This fold is a common topology for enzymes and can harbor many diverse reactions. The recent de novo design of a four-fold symmetric TIM barrel provides a well understood minimal scaffold for potential enzyme designs. Here we explore opportunities to extend and diversify this scaffold by adding a short de novo helix on top of the barrel. Due to the size of the protein, we developed a design pipeline based on computational ab initio folding that solves a less complex sub-problem focused around the helix and its vicinity and adapt it to the entire protein. We provide biochemical characterization and a high-resolution X-ray structure for one variant and compare it to our design model. The successful extension of this robust TIM-barrel scaffold opens opportunities to diversify it towards more pocket like arrangements and as such can be considered a building block for future design of binding or catalytic sites.
Organizational Affiliation:
Max Planck Institute for Developmental Biology, Tübingen, Germany.