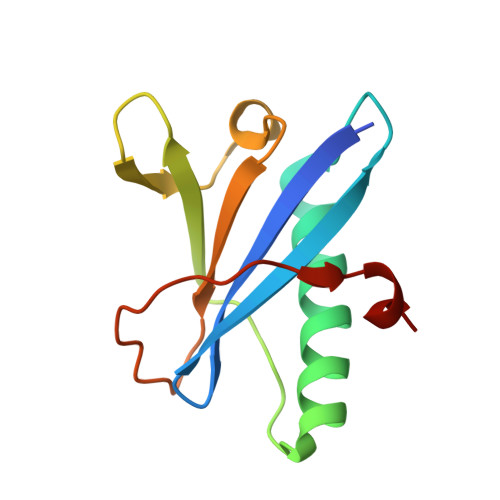
Structural and functional studies of the first tripartite protein complex at the Trypanosoma brucei flagellar pocket collar.
Isch, C., Majneri, P., Landrein, N., Pivovarova, Y., Lesigang, J., Lauruol, F., Robinson, D.R., Dong, G., Bonhivers, M.(2021) PLoS Pathog 17: e1009329-e1009329
- PubMed: 34339455
- DOI: https://doi.org/10.1371/journal.ppat.1009329
- Primary Citation of Related Structures:
7A1I - PubMed Abstract:
The flagellar pocket (FP) is the only endo- and exocytic organelle in most trypanosomes and, as such, is essential throughout the life cycle of the parasite. The neck of the FP is maintained enclosed around the flagellum via the flagellar pocket collar (FPC). The FPC is a macromolecular cytoskeletal structure and is essential for the formation of the FP and cytokinesis. FPC biogenesis and structure are poorly understood, mainly due to the lack of information on FPC composition. To date, only two FPC proteins, BILBO1 and FPC4, have been characterized. BILBO1 forms a molecular skeleton upon which other FPC proteins can, theoretically, dock onto. We previously identified FPC4 as the first BILBO1 interacting partner and demonstrated that its C-terminal domain interacts with the BILBO1 N-terminal domain (NTD). Here, we report by yeast two-hybrid, bioinformatics, functional and structural studies the characterization of a new FPC component and BILBO1 partner protein, BILBO2 (Tb927.6.3240). Further, we demonstrate that BILBO1 and BILBO2 share a homologous NTD and that both domains interact with FPC4. We have determined a 1.9 Å resolution crystal structure of the BILBO2 NTD in complex with the FPC4 BILBO1-binding domain. Together with mutational analyses, our studies reveal key residues for the function of the BILBO2 NTD and its interaction with FPC4 and evidenced a tripartite interaction between BILBO1, BILBO2, and FPC4. Our work sheds light on the first atomic structure of an FPC protein complex and represents a significant step in deciphering the FPC function in Trypanosoma brucei and other pathogenic kinetoplastids.
Organizational Affiliation:
Univ. Bordeaux, CNRS, Microbiologie Fondamentale et Pathogénicité, UMR 5234, Bordeaux, France.