Crystal structure of Pseudomonas aeruginosa S-adenosyl-L-homocysteine hydrolase inhibited by Zn2+ ions
Malecki, P.H., Gawel, M., Brzezinski, K.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Adenosylhomocysteinase | 472 | Pseudomonas aeruginosa PAO1 | Mutation(s): 0 Gene Names: ahcY, sahH, PA0432 EC: 3.3.1.1 | 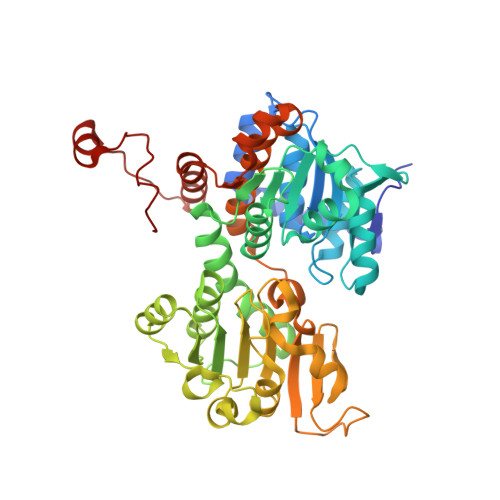 | |
UniProt | |||||
Find proteins for Q9I685 (Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1)) Explore Q9I685 Go to UniProtKB: Q9I685 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9I685 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 10 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAD (Subject of Investigation/LOI) Query on NAD | AA [auth C], E [auth A], KA [auth D], P [auth B] | NICOTINAMIDE-ADENINE-DINUCLEOTIDE C21 H27 N7 O14 P2 BAWFJGJZGIEFAR-NNYOXOHSSA-N |  | ||
| ADN (Subject of Investigation/LOI) Query on ADN | BA [auth C], F [auth A], LA [auth D], Q [auth B] | ADENOSINE C10 H13 N5 O4 OIRDTQYFTABQOQ-KQYNXXCUSA-N |  | ||
| PG4 Query on PG4 | I [auth A] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| BCN Query on BCN | Y [auth B] | BICINE C6 H13 N O4 FSVCELGFZIQNCK-UHFFFAOYSA-N |  | ||
| HEZ Query on HEZ | EA [auth C], Z [auth C] | HEXANE-1,6-DIOL C6 H14 O2 XXMIOPMDWAUFGU-UHFFFAOYSA-N |  | ||
| BU1 Query on BU1 | G [auth A], H [auth A], J [auth A], R [auth B], S [auth B] | 1,4-BUTANEDIOL C4 H10 O2 WERYXYBDKMZEQL-UHFFFAOYSA-N |  | ||
| PDO Query on PDO | CA [auth C], DA [auth C], FA [auth C] | 1,3-PROPANDIOL C3 H8 O2 YPFDHNVEDLHUCE-UHFFFAOYSA-N |  | ||
| ZN (Subject of Investigation/LOI) Query on ZN | HA [auth C] IA [auth C] JA [auth C] L [auth A] M [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| K (Subject of Investigation/LOI) Query on K | GA [auth C], K [auth A], MA [auth D], T [auth B] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| CL Query on CL | SA [auth D] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 177.19 | α = 90 |
| b = 134.05 | β = 105.705 |
| c = 108.46 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| REFMAC | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Polish National Science Centre | Poland | SONATA BIS 2018/30/E/NZ1/00729 |