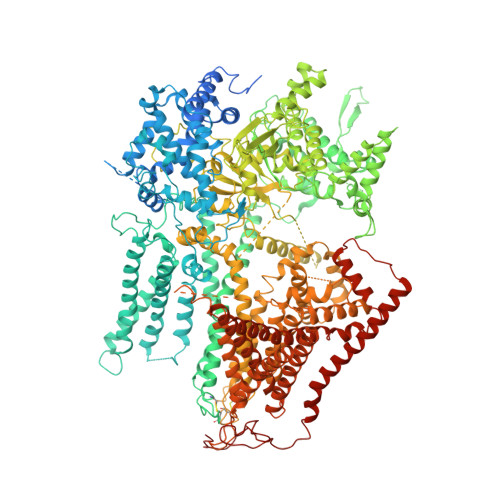
Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Hu, X., Yang, P., Chai, C., Liu, J., Sun, H., Wu, Y., Zhang, M., Zhang, M., Liu, X., Yu, H.(2023) Nature 616: 190-198
- PubMed: 36949198
- DOI: https://doi.org/10.1038/s41586-023-05856-5
- Primary Citation of Related Structures:
7XE4, 7YUY - PubMed Abstract:
The membrane-integrated synthase FKS is involved in the biosynthesis of β-1,3-glucan, the core component of the fungal cell wall 1,2 . FKS is the target of widely prescribed antifungal drugs, including echinocandin and ibrexafungerp 3,4 . Unfortunately, the mechanism of action of FKS remains enigmatic and this has hampered development of more effective medicines targeting the enzyme. Here we present the cryo-electron microscopy structures of Saccharomyces cerevisiae FKS1 and the echinocandin-resistant mutant FKS1(S643P). These structures reveal the active site of the enzyme at the membrane-cytoplasm interface and a glucan translocation path spanning the membrane bilayer. Multiple bound lipids and notable membrane distortions are observed in the FKS1 structures, suggesting active FKS1-membrane interactions. Echinocandin-resistant mutations are clustered at a region near TM5-6 and TM8 of FKS1. The structure of FKS1(S643P) reveals altered lipid arrangements in this region, suggesting a drug-resistant mechanism of the mutant enzyme. The structures, the catalytic mechanism and the molecular insights into drug-resistant mutations of FKS1 revealed in this study advance the mechanistic understanding of fungal β-1,3-glucan biosynthesis and establish a foundation for developing new antifungal drugs by targeting FKS.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, School of Basic Medicine, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.