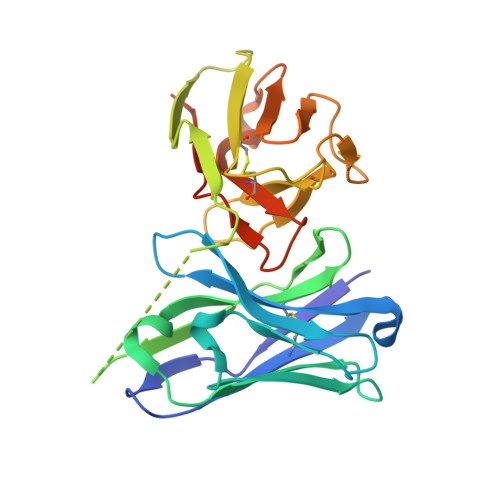
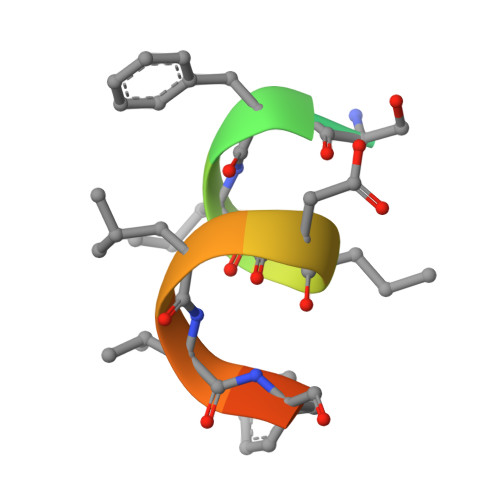
Epitope-directed anti-SARS-CoV-2 scFv engineered against the key spike protein region could block membrane fusion.
Jaiswal, D., Kumar, U., Gaur, V., Salunke, D.M.(2023) Protein Sci 32: e4575-e4575
- PubMed: 36691733
- DOI: https://doi.org/10.1002/pro.4575
- Primary Citation of Related Structures:
7YUE - PubMed Abstract:
The newly emerged SARS-CoV-2 causing coronavirus disease (COVID-19) resulted in >500 million infections. A great deal about the molecular processes of virus infection in the host is getting uncovered. Two sequential proteolytic cleavages of viral spike protein by host proteases are prerequisites for the entry of the virus into the host cell. The first cleavage occurs at S1/S2 site by the furin protease, and the second cleavage at a fusion activation site, the S2' site, by the TMPRSS2 protease. S2' cleavage site is present in the S2 domain of spike protein followed by a fusion peptide. Given the S2' site to be conserved among all the SARS-CoV-2 variants, we chose an S2' epitope encompassing the S2' cleavage site and generated single-chain antibodies (scFvs) through an exhaustive phage display library screening. Crystal structure of a scFv in complex with S2' epitope was determined. Incidentally, S2' epitope in the scFv bound structure adopts an alpha-helical conformation equivalent to the conformation of the epitope in the spike protein. Furthermore, these scFvs can bind to the spike protein expressed either in vitro or on the mammalian cell surface. We illustrate a molecular model based on structural and biochemical insights into the antibody-S2' epitope interaction emphasizing scFvs mediated blocking of virus entry into the host cell by restricting the access of TMPRSS2 protease and consequently inhibiting the S2' cleavage competitively.
Organizational Affiliation:
International Centre for Genetic Engineering and Biotechnology, New Delhi, Delhi, India.