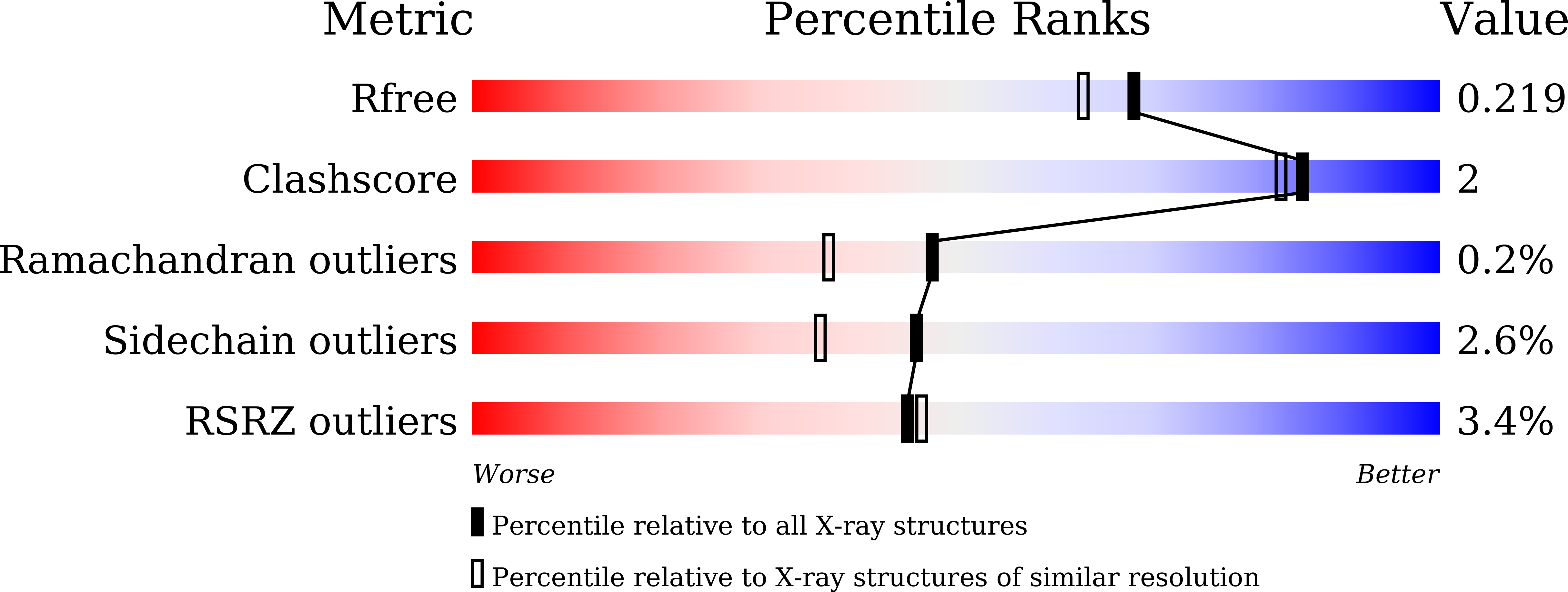
Biochemical and crystallographic assessments of the effect of 4,6-O-disulfated disaccharide moieties in chondroitin sulfate E on chondroitinase ABC I activity.
Watanabe, I., Miyanaga, A., Hoshi, H., Suzuki, K., Eguchi, T.(2023) FEBS J 290: 2379-2393
- PubMed: 36478634
- DOI: https://doi.org/10.1111/febs.16685
- Primary Citation of Related Structures:
7YKE - PubMed Abstract:
O-sulfated N-acetyl-d-galactosamine (GalNAc) residues in chondroitin sulfate (CS) play a crucial role in chondroitinase ABC I (cABC-I) activity. CSA containing mainly 4-O-monosulfated GalNAc was a good substrate for the enzyme, but not CSE containing mainly 4,6-O-disulfated GalNAc [GalNAc(4S,6S)]. Each CS isomer exhibits structural heterogeneity; CSE has di-sulfated disaccharide units and mono-sulfated disaccharide units. Disaccharide composition analysis of digested products revealed that mono-sulfated disaccharide units in CSE contributed to the enzyme reactivity. Although enough substrate (CSA) was present in mixtures of CSA and CSE for reaction, the reactivity was reduced depending on the amount of CSE in the mixture. These results suggested that CSE is not only resistant to enzyme digestion but also attenuates enzyme activity. To understand the mechanism of action, crystallography of cABC-I in complex with unsaturated CSE-disaccharide, ΔDi-(4,6)S, was performed. Both 4-O- and 6-O-sulfate groups in ΔDi-(4,6)S interact with Arg500, suggesting that there was a greater interaction between ΔDi-(4,6)S and Arg500 than between mono-sulfated disaccharides and Arg500. Besides, this interaction attenuated enzyme activity by interfering with a function of Arg500, which is the charge neutralization of the carboxy group of D-glucuronic acid (GlcA) residues in CS. When interacting with the CSE-disaccharide unit [GlcAβ1-3GalNAc(4S,6S)] in CS, cABC-I cannot interact with other CS-disaccharide units until it has digested the CSE-disaccharide unit. The low reactivity of cABC-I with CSE is attributable to two suggested factors: (a) resistance of E-units in CSE molecules to digestion by cABC-I, and (b) tendency of E-units in CSE molecules to attenuate cABC-I activity.
Organizational Affiliation:
Medical Affairs, Seikagaku Corporation, Chiyoda-ku, Japan.