Functional analysis of the N-terminal region of acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis.
Sasamoto, K., Himiyama, T., Moriyoshi, K., Ohmoto, T., Uegaki, K., Nakamura, T., Nishiya, Y.(2022) FEBS Open Bio 12: 1875-1885
- PubMed: 36054591
- DOI: https://doi.org/10.1002/2211-5463.13476
- Primary Citation of Related Structures:
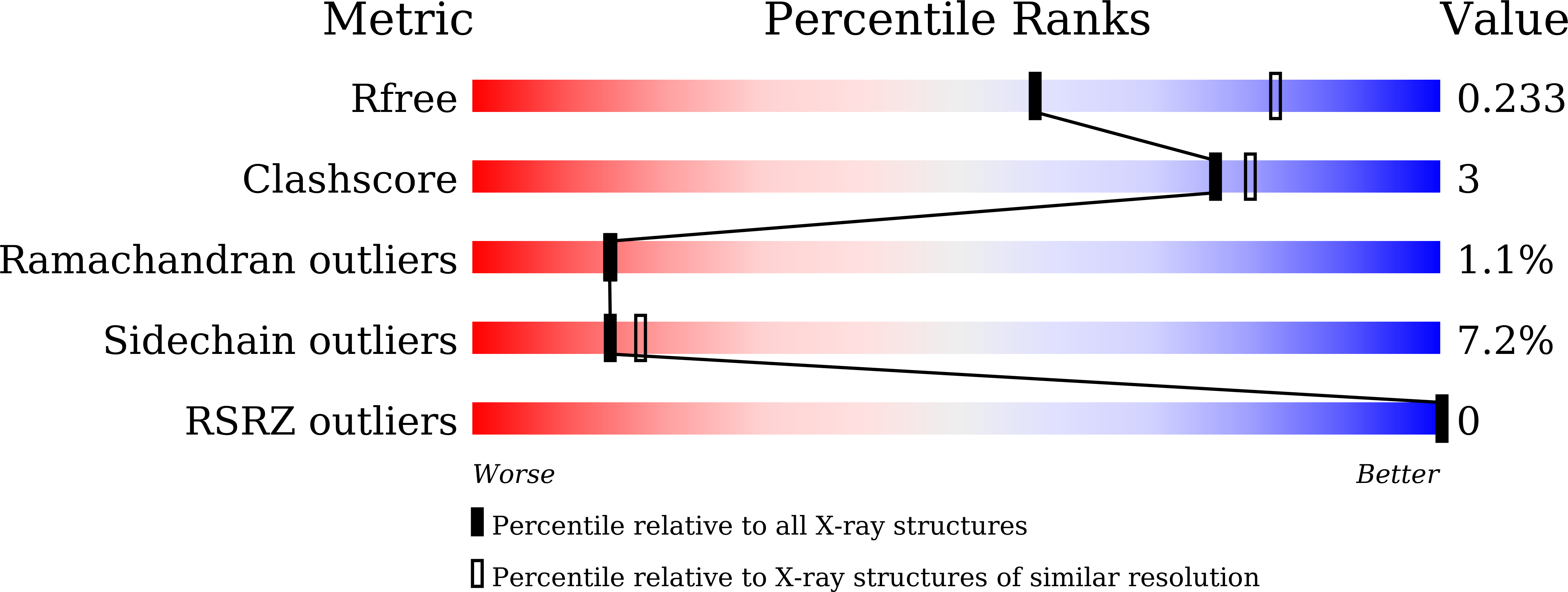
7Y51 - PubMed Abstract:
Acetylxylan esterase from Caldanaerobacter subterraneus subsp. tengcongensis (TTE0866) has an N-terminal region (NTR; residues 23-135) between the signal sequence (residues 1-22) and the catalytic domain (residues 136-324), which is of unknown function. Our previous study revealed the crystal structure of the wild-type (WT) enzyme containing the NTR and the catalytic domain. Although the structure of the catalytic domain was successfully determined, that of the NTR was undetermined, as its electron density was unclear. In this study, we investigated the role of the NTR through functional and structural analyses of NTR truncation mutants. Based on sequence and secondary structure analyses, NTR was confirmed to be an intrinsically disordered region. The truncation of NTR significantly decreased the solubility of the proteins at low salt concentrations compared with that of the WT. The NTR-truncated mutant easily crystallized in a conventional buffer solution. The crystal exhibited crystallographic properties comparable with those of the WT crystals suitable for structural determination. These results suggest that NTR plays a role in maintaining the solubility and inhibiting the crystallization of the catalytic domain.
Organizational Affiliation:
Division of Life Science, Graduate School of Science and Engineering, Setsunan University, Osaka, Japan.