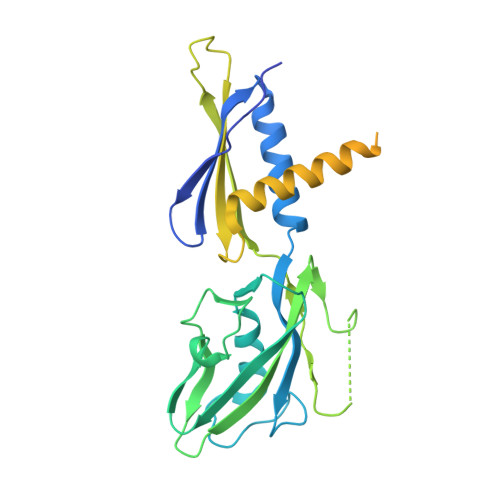
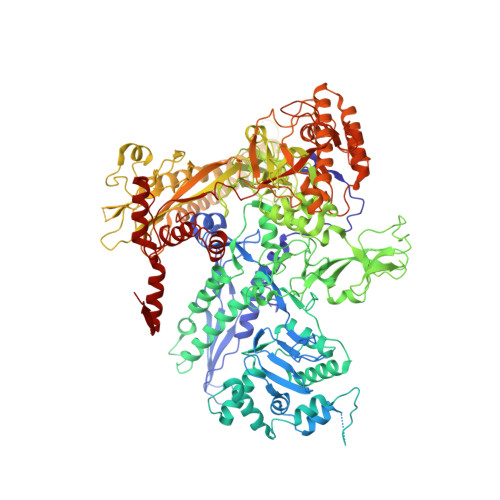
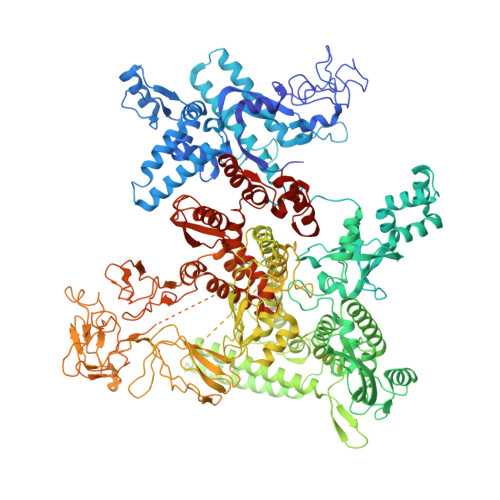
Structural basis of AlpA-dependent transcription antitermination.
Wen, A., Zhao, M., Jin, S., Lu, Y.Q., Feng, Y.(2022) Nucleic Acids Res 50: 8321-8330
- PubMed: 35871295
- DOI: https://doi.org/10.1093/nar/gkac608
- Primary Citation of Related Structures:
7XYA, 7XYB - PubMed Abstract:
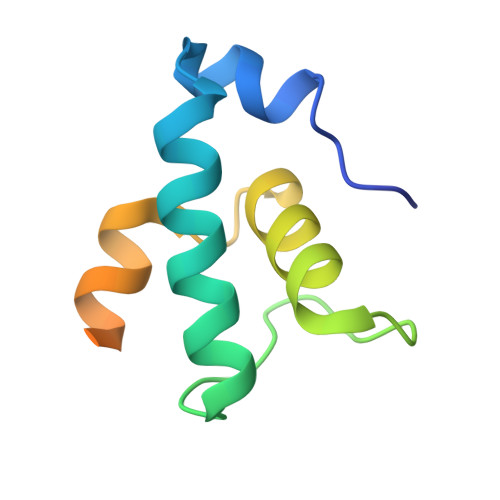
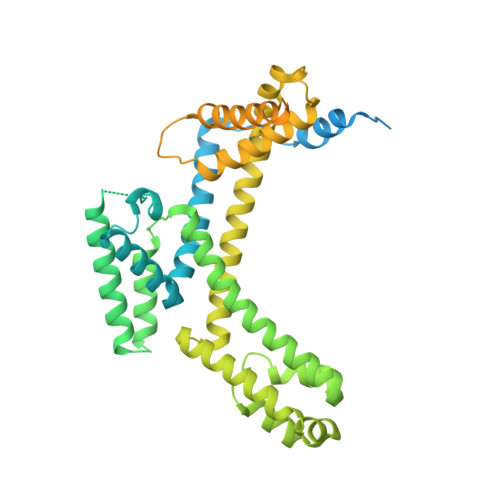
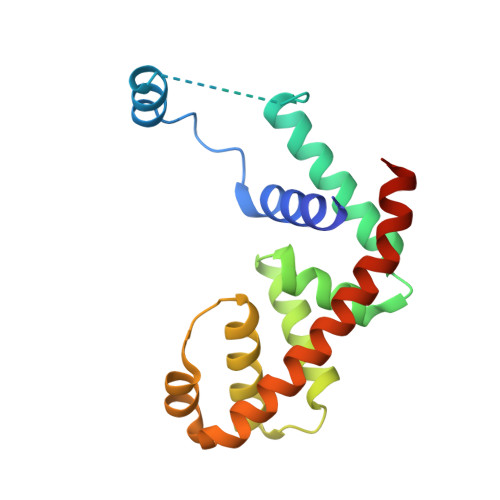
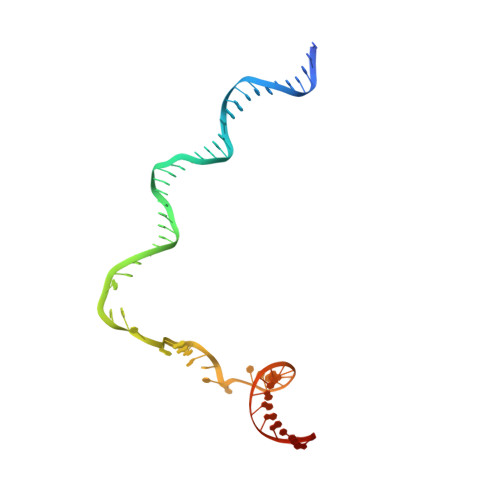
AlpA positively regulates a programmed cell death pathway linked to the virulence of Pseudomonas aeruginosa by recognizing an AlpA binding element within the promoter, then binding RNA polymerase directly and allowing it to bypass an intrinsic terminator positioned downstream. Here, we report the single-particle cryo-electron microscopy structures of both an AlpA-loading complex and an AlpA-loaded complex. These structures indicate that the C-terminal helix-turn-helix motif of AlpA binds to the AlpA binding element and that the N-terminal segment of AlpA forms a narrow ring inside the RNA exit channel. AlpA was also revealed to render RNAP resistant to termination signals by prohibiting RNA hairpin formation in the RNA exit channel. Structural analysis predicted that AlpA, 21Q, λQ and 82Q share the same mechanism of transcription antitermination.
Organizational Affiliation:
Department of Biophysics, and Department of Infectious Disease of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou 310058, China.