Parallel T Cell Immunogenic Regions in Influenza B and A Viruses with Distinct Nuclear Export Signal Functions: The Balance between Viral Life Cycle and Immune Escape.
Zhao, Y., Xiao, W., Wu, Y., Fan, W., Li, L., Yue, C., Zhang, Q., Zhang, D., Yuan, X., Yao, S., Liu, S., Li, M., Wang, P., Zhang, H., Zhang, J., Zhao, M., Zheng, X., Liu, W., Gao, G.F., Liu, W.J.(2023) J Immunol 210: 1074-1085
- PubMed: 36897229
- DOI: https://doi.org/10.4049/jimmunol.2200243
- Primary Citation of Related Structures:
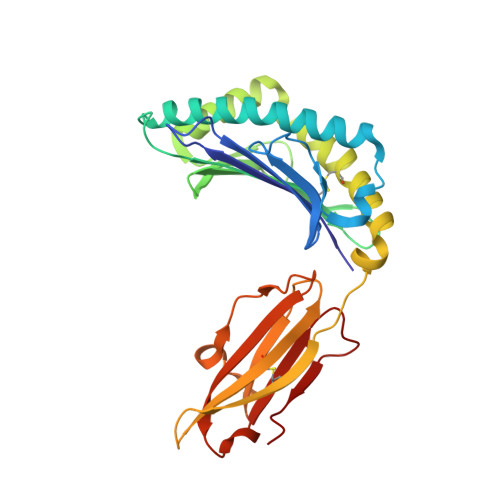
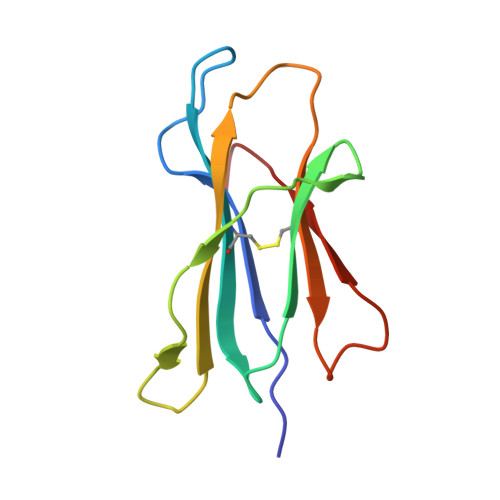
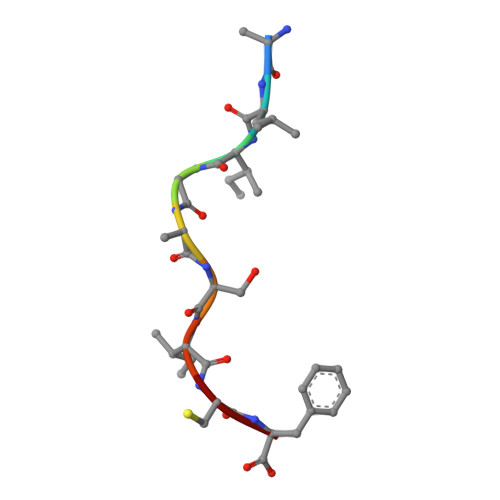
7XF3 - PubMed Abstract:
Influenza A viruses (IAVs) and influenza B viruses (IBVs) cause annual epidemics in human populations with seasonal circulation spikes. Peptide AM58-66GL9 located at residues 58-66 of M1 protein of IAVs has been recognized as an immunodominant T cell epitope with HLA-A*0201 restriction and broadly used as a positive reference in influenza immunity. This peptide also almost completely overlaps with a nuclear export signal (NES) 59-68 in IAV M1, which explains the limited escape mutations under the T cell immune pressure in this region. In this study, we investigated the potential immunogenicity and NES in the corresponding region of IBV. The long peptide covering this region can be recognized by specific T cells and induce robust expression of IFN-γ among HLA-B*1501 donors in vivo, but not in HLA-A*0201 donors. Among a series of truncated peptides derived from this region, we identified an immunodominant HLA-B*1501-restricted T cell epitope BM58-66AF9 (ALIGASICF) in the M1 protein of IBV. Furthermore, the structure of the HLA-B*1501/BM58-66AF9 complex shows that BM58-66AF9 performs a flat and featureless conformation that is similar to AM58-66GL9 presented by HLA-A*0201. In contrast with IAV, the sequence around residues 55-70 of IBV M1 does not contain an NES. Our comparative study on IBVs and IAVs provides new insights into the immune and evolution characteristics of IBVs and may shed light on vaccine development for influenza viruses.
Organizational Affiliation:
NHC Key Laboratory of Biosafety, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China.